Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base

Multiple Linear Regression | A Quick Guide (Examples)
Published on February 20, 2020 by Rebecca Bevans . Revised on June 22, 2023.
Regression models are used to describe relationships between variables by fitting a line to the observed data. Regression allows you to estimate how a dependent variable changes as the independent variable(s) change.
Multiple linear regression is used to estimate the relationship between two or more independent variables and one dependent variable . You can use multiple linear regression when you want to know:
- How strong the relationship is between two or more independent variables and one dependent variable (e.g. how rainfall, temperature, and amount of fertilizer added affect crop growth).
- The value of the dependent variable at a certain value of the independent variables (e.g. the expected yield of a crop at certain levels of rainfall, temperature, and fertilizer addition).
Table of contents
Assumptions of multiple linear regression, how to perform a multiple linear regression, interpreting the results, presenting the results, other interesting articles, frequently asked questions about multiple linear regression.
Multiple linear regression makes all of the same assumptions as simple linear regression :
Homogeneity of variance (homoscedasticity) : the size of the error in our prediction doesn’t change significantly across the values of the independent variable.
Independence of observations : the observations in the dataset were collected using statistically valid sampling methods , and there are no hidden relationships among variables.
In multiple linear regression, it is possible that some of the independent variables are actually correlated with one another, so it is important to check these before developing the regression model. If two independent variables are too highly correlated (r2 > ~0.6), then only one of them should be used in the regression model.
Normality : The data follows a normal distribution .
Linearity : the line of best fit through the data points is a straight line, rather than a curve or some sort of grouping factor.
Here's why students love Scribbr's proofreading services
Discover proofreading & editing
Multiple linear regression formula
The formula for a multiple linear regression is:
- … = do the same for however many independent variables you are testing
To find the best-fit line for each independent variable, multiple linear regression calculates three things:
- The regression coefficients that lead to the smallest overall model error.
- The t statistic of the overall model.
- The associated p value (how likely it is that the t statistic would have occurred by chance if the null hypothesis of no relationship between the independent and dependent variables was true).
It then calculates the t statistic and p value for each regression coefficient in the model.
Multiple linear regression in R
While it is possible to do multiple linear regression by hand, it is much more commonly done via statistical software. We are going to use R for our examples because it is free, powerful, and widely available. Download the sample dataset to try it yourself.
Dataset for multiple linear regression (.csv)
Load the heart.data dataset into your R environment and run the following code:
This code takes the data set heart.data and calculates the effect that the independent variables biking and smoking have on the dependent variable heart disease using the equation for the linear model: lm() .
Learn more by following the full step-by-step guide to linear regression in R .
To view the results of the model, you can use the summary() function:
This function takes the most important parameters from the linear model and puts them into a table that looks like this:
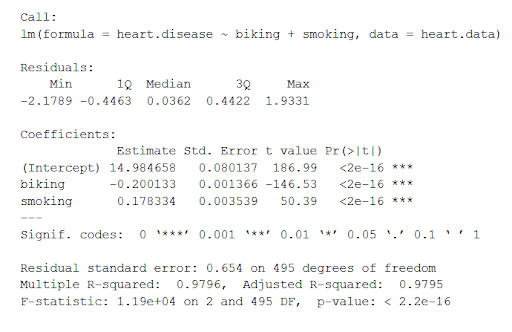
The summary first prints out the formula (‘Call’), then the model residuals (‘Residuals’). If the residuals are roughly centered around zero and with similar spread on either side, as these do ( median 0.03, and min and max around -2 and 2) then the model probably fits the assumption of heteroscedasticity.
Next are the regression coefficients of the model (‘Coefficients’). Row 1 of the coefficients table is labeled (Intercept) – this is the y-intercept of the regression equation. It’s helpful to know the estimated intercept in order to plug it into the regression equation and predict values of the dependent variable:
The most important things to note in this output table are the next two tables – the estimates for the independent variables.
The Estimate column is the estimated effect , also called the regression coefficient or r 2 value. The estimates in the table tell us that for every one percent increase in biking to work there is an associated 0.2 percent decrease in heart disease, and that for every one percent increase in smoking there is an associated .17 percent increase in heart disease.
The Std.error column displays the standard error of the estimate. This number shows how much variation there is around the estimates of the regression coefficient.
The t value column displays the test statistic . Unless otherwise specified, the test statistic used in linear regression is the t value from a two-sided t test . The larger the test statistic, the less likely it is that the results occurred by chance.
The Pr( > | t | ) column shows the p value . This shows how likely the calculated t value would have occurred by chance if the null hypothesis of no effect of the parameter were true.
Because these values are so low ( p < 0.001 in both cases), we can reject the null hypothesis and conclude that both biking to work and smoking both likely influence rates of heart disease.
When reporting your results, include the estimated effect (i.e. the regression coefficient), the standard error of the estimate, and the p value. You should also interpret your numbers to make it clear to your readers what the regression coefficient means.
Visualizing the results in a graph
It can also be helpful to include a graph with your results. Multiple linear regression is somewhat more complicated than simple linear regression, because there are more parameters than will fit on a two-dimensional plot.
However, there are ways to display your results that include the effects of multiple independent variables on the dependent variable, even though only one independent variable can actually be plotted on the x-axis.
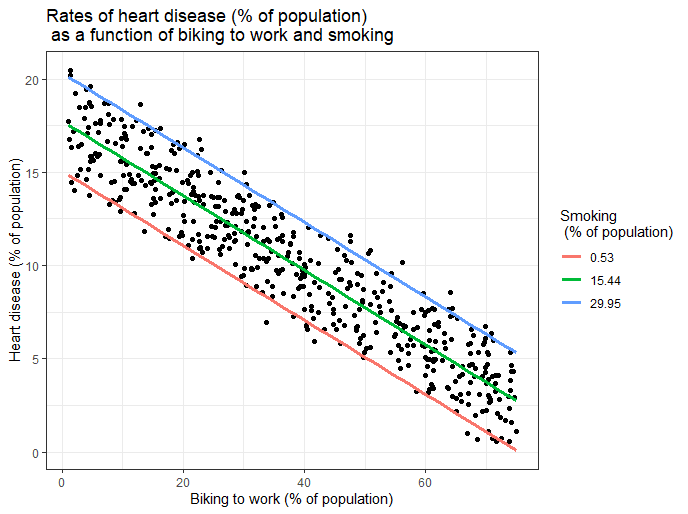
Here, we have calculated the predicted values of the dependent variable (heart disease) across the full range of observed values for the percentage of people biking to work.
To include the effect of smoking on the independent variable, we calculated these predicted values while holding smoking constant at the minimum, mean , and maximum observed rates of smoking.
Receive feedback on language, structure, and formatting
Professional editors proofread and edit your paper by focusing on:
- Academic style
- Vague sentences
- Style consistency
See an example
If you want to know more about statistics , methodology , or research bias , make sure to check out some of our other articles with explanations and examples.
- Chi square test of independence
- Statistical power
- Descriptive statistics
- Degrees of freedom
- Pearson correlation
- Null hypothesis
Methodology
- Double-blind study
- Case-control study
- Research ethics
- Data collection
- Hypothesis testing
- Structured interviews
Research bias
- Hawthorne effect
- Unconscious bias
- Recall bias
- Halo effect
- Self-serving bias
- Information bias
A regression model is a statistical model that estimates the relationship between one dependent variable and one or more independent variables using a line (or a plane in the case of two or more independent variables).
A regression model can be used when the dependent variable is quantitative, except in the case of logistic regression, where the dependent variable is binary.
Multiple linear regression is a regression model that estimates the relationship between a quantitative dependent variable and two or more independent variables using a straight line.
Linear regression most often uses mean-square error (MSE) to calculate the error of the model. MSE is calculated by:
- measuring the distance of the observed y-values from the predicted y-values at each value of x;
- squaring each of these distances;
- calculating the mean of each of the squared distances.
Linear regression fits a line to the data by finding the regression coefficient that results in the smallest MSE.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the “Cite this Scribbr article” button to automatically add the citation to our free Citation Generator.
Bevans, R. (2023, June 22). Multiple Linear Regression | A Quick Guide (Examples). Scribbr. Retrieved September 5, 2024, from https://www.scribbr.com/statistics/multiple-linear-regression/
Is this article helpful?
Rebecca Bevans
Other students also liked, simple linear regression | an easy introduction & examples, an introduction to t tests | definitions, formula and examples, types of variables in research & statistics | examples, what is your plagiarism score.
Multiple linear regression
Multiple linear regression #.
Fig. 11 Multiple linear regression #
Errors: \(\varepsilon_i \sim N(0,\sigma^2)\quad \text{i.i.d.}\)
Fit: the estimates \(\hat\beta_0\) and \(\hat\beta_1\) are chosen to minimize the residual sum of squares (RSS):
Matrix notation: with \(\beta=(\beta_0,\dots,\beta_p)\) and \({X}\) our usual data matrix with an extra column of ones on the left to account for the intercept, we can write
Multiple linear regression answers several questions #
Is at least one of the variables \(X_i\) useful for predicting the outcome \(Y\) ?
Which subset of the predictors is most important?
How good is a linear model for these data?
Given a set of predictor values, what is a likely value for \(Y\) , and how accurate is this prediction?
The estimates \(\hat\beta\) #
Our goal again is to minimize the RSS: $ \( \begin{aligned} \text{RSS}(\beta) &= \sum_{i=1}^n (y_i -\hat y_i(\beta))^2 \\ & = \sum_{i=1}^n (y_i - \beta_0- \beta_1 x_{i,1}-\dots-\beta_p x_{i,p})^2 \\ &= \|Y-X\beta\|^2_2 \end{aligned} \) $
One can show that this is minimized by the vector \(\hat\beta\) : $ \(\hat\beta = ({X}^T{X})^{-1}{X}^T{y}.\) $
We usually write \(RSS=RSS(\hat{\beta})\) for the minimized RSS.
Which variables are important? #
Consider the hypothesis: \(H_0:\) the last \(q\) predictors have no relation with \(Y\) .
Based on our model: \(H_0:\beta_{p-q+1}=\beta_{p-q+2}=\dots=\beta_p=0.\)
Let \(\text{RSS}_0\) be the minimized residual sum of squares for the model which excludes these variables.
The \(F\) -statistic is defined by: $ \(F = \frac{(\text{RSS}_0-\text{RSS})/q}{\text{RSS}/(n-p-1)}.\) $
Under the null hypothesis (of our model), this has an \(F\) -distribution.
Example: If \(q=p\) , we test whether any of the variables is important. $ \(\text{RSS}_0 = \sum_{i=1}^n(y_i-\overline y)^2 \) $
| Res.Df | RSS | Df | Sum of Sq | F | Pr(>F) |
|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> |
| 494 | 11336.29 | NA | NA | NA | NA |
| 492 | 11078.78 | 2 | 257.5076 | 5.717853 | 0.003509036 |
The \(t\) -statistic associated to the \(i\) th predictor is the square root of the \(F\) -statistic for the null hypothesis which sets only \(\beta_i=0\) .
A low \(p\) -value indicates that the predictor is important.
Warning: If there are many predictors, even under the null hypothesis, some of the \(t\) -tests will have low p-values even when the model has no explanatory power.
How many variables are important? #
When we select a subset of the predictors, we have \(2^p\) choices.
A way to simplify the choice is to define a range of models with an increasing number of variables, then select the best.
Forward selection: Starting from a null model, include variables one at a time, minimizing the RSS at each step.
Backward selection: Starting from the full model, eliminate variables one at a time, choosing the one with the largest p-value at each step.
Mixed selection: Starting from some model, include variables one at a time, minimizing the RSS at each step. If the p-value for some variable goes beyond a threshold, eliminate that variable.
Choosing one model in the range produced is a form of tuning . This tuning can invalidate some of our methods like hypothesis tests and confidence intervals…
How good are the predictions? #
The function predict in R outputs predictions and confidence intervals from a linear model:
| fit | lwr | upr |
|---|---|---|
| 9.409426 | 8.722696 | 10.09616 |
| 14.163090 | 13.708423 | 14.61776 |
| 18.916754 | 18.206189 | 19.62732 |
Prediction intervals reflect uncertainty on \(\hat\beta\) and the irreducible error \(\varepsilon\) as well.
| fit | lwr | upr |
|---|---|---|
| 9.409426 | 2.946709 | 15.87214 |
| 14.163090 | 7.720898 | 20.60528 |
| 18.916754 | 12.451461 | 25.38205 |
These functions rely on our linear regression model $ \( Y = X\beta + \epsilon. \) $
Dealing with categorical or qualitative predictors #
For each qualitative predictor, e.g. Region :
Choose a baseline category, e.g. East
For every other category, define a new predictor:
\(X_\text{South}\) is 1 if the person is from the South region and 0 otherwise
\(X_\text{West}\) is 1 if the person is from the West region and 0 otherwise.
The model will be: $ \(Y = \beta_0 + \beta_1 X_1 +\dots +\beta_7 X_7 + \color{Red}{\beta_\text{South}} X_\text{South} + \beta_\text{West} X_\text{West} +\varepsilon.\) $
The parameter \(\color{Red}{\beta_\text{South}}\) is the relative effect on Balance (our \(Y\) ) for being from the South compared to the baseline category (East).
The model fit and predictions are independent of the choice of the baseline category.
However, hypothesis tests derived from these variables are affected by the choice.
Solution: To check whether region is important, use an \(F\) -test for the hypothesis \(\beta_\text{South}=\beta_\text{West}=0\) by dropping Region from the model. This does not depend on the coding.
Note that there are other ways to encode qualitative predictors produce the same fit \(\hat f\) , but the coefficients have different interpretations.
So far, we have:
Defined Multiple Linear Regression
Discussed how to test the importance of variables.
Described one approach to choose a subset of variables.
Explained how to code qualitative variables.
Now, how do we evaluate model fit? Is the linear model any good? What can go wrong?
How good is the fit? #
To assess the fit, we focus on the residuals $ \( e = Y - \hat{Y} \) $
The RSS always decreases as we add more variables.
The residual standard error (RSE) corrects this: $ \(\text{RSE} = \sqrt{\frac{1}{n-p-1}\text{RSS}}.\) $
Fig. 12 Residuals #
Visualizing the residuals can reveal phenomena that are not accounted for by the model; eg. synergies or interactions:
Potential issues in linear regression #
Interactions between predictors
Non-linear relationships
Correlation of error terms
Non-constant variance of error (heteroskedasticity)
High leverage points
Collinearity
Interactions between predictors #
Linear regression has an additive assumption: $ \(\mathtt{sales} = \beta_0 + \beta_1\times\mathtt{tv}+ \beta_2\times\mathtt{radio}+\varepsilon\) $
i.e. An increase of 100 USD dollars in TV ads causes a fixed increase of \(100 \beta_2\) USD in sales on average, regardless of how much you spend on radio ads.
We saw that in Fig 3.5 above. If we visualize the fit and the observed points, we see they are not evenly scattered around the plane. This could be caused by an interaction.
One way to deal with this is to include multiplicative variables in the model:
The interaction variable tv \(\cdot\) radio is high when both tv and radio are high.
R makes it easy to include interaction variables in the model:
Non-linearities #
Fig. 13 A nonlinear fit might be better here. #
Example: Auto dataset.
A scatterplot between a predictor and the response may reveal a non-linear relationship.
Solution: include polynomial terms in the model.
Could use other functions besides polynomials…
Fig. 14 Residuals for Auto data #
In 2 or 3 dimensions, this is easy to visualize. What do we do when we have too many predictors?
Correlation of error terms #
We assumed that the errors for each sample are independent:
What if this breaks down?
The main effect is that this invalidates any assertions about Standard Errors, confidence intervals, and hypothesis tests…
Example : Suppose that by accident, we duplicate the data (we use each sample twice). Then, the standard errors would be artificially smaller by a factor of \(\sqrt{2}\) .
When could this happen in real life:
Time series: Each sample corresponds to a different point in time. The errors for samples that are close in time are correlated.
Spatial data: Each sample corresponds to a different location in space.
Grouped data: Imagine a study on predicting height from weight at birth. If some of the subjects in the study are in the same family, their shared environment could make them deviate from \(f(x)\) in similar ways.
Correlated errors #
Simulations of time series with increasing correlations between \(\varepsilon_i\)
Non-constant variance of error (heteroskedasticity) #
The variance of the error depends on some characteristics of the input features.
To diagnose this, we can plot residuals vs. fitted values:
If the trend in variance is relatively simple, we can transform the response using a logarithm, for example.
Outliers from a model are points with very high errors.
While they may not affect the fit, they might affect our assessment of model quality.
Possible solutions: #
If we believe an outlier is due to an error in data collection, we can remove it.
An outlier might be evidence of a missing predictor, or the need to specify a more complex model.
High leverage points #
Some samples with extreme inputs have an outsized effect on \(\hat \beta\) .
This can be measured with the leverage statistic or self influence :
Studentized residuals #
The residual \(e_i = y_i - \hat y_i\) is an estimate for the noise \(\epsilon_i\) .
The standard error of \(\hat \epsilon_i\) is \(\sigma \sqrt{1-h_{ii}}\) .
A studentized residual is \(\hat \epsilon_i\) divided by its standard error (with appropriate estimate of \(\sigma\) )
When model is correct, it follows a Student-t distribution with \(n-p-2\) degrees of freedom.
Collinearity #
Two predictors are collinear if one explains the other well:
Problem: The coefficients become unidentifiable .
Consider the extreme case of using two identical predictors limit : $ \( \begin{aligned} \mathtt{balance} &= \beta_0 + \beta_1\times\mathtt{limit} + \beta_2\times\mathtt{limit} + \epsilon \\ & = \beta_0 + (\beta_1+100)\times\mathtt{limit} + (\beta_2-100)\times\mathtt{limit} + \epsilon \end{aligned} \) $
For every \((\beta_0,\beta_1,\beta_2)\) the fit at \((\beta_0,\beta_1,\beta_2)\) is just as good as at \((\beta_0,\beta_1+100,\beta_2-100)\) .
If 2 variables are collinear, we can easily diagnose this using their correlation.
A group of \(q\) variables is multilinear if these variables “contain less information” than \(q\) independent variables.
Pairwise correlations may not reveal multilinear variables.
The Variance Inflation Factor (VIF) measures how predictable it is given the other variables, a proxy for how necessary a variable is:
Above, \(R^2_{X_j|X_{-j}}\) is the \(R^2\) statistic for Multiple Linear regression of the predictor \(X_j\) onto the remaining predictors.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Hippokratia
- v.14(Suppl 1); 2010 Dec
Introduction to Multivariate Regression Analysis
Statistics are used in medicine for data description and inference. Inferential statistics are used to answer questions about the data, to test hypotheses (formulating the alternative or null hypotheses), to generate a measure of effect, typically a ratio of rates or risks, to describe associations (correlations) or to model relationships (regression) within the data and, in many other functions. Usually point estimates are the measures of associations or of the magnitude of effects. Confounding, measurement errors, selection bias and random errors make unlikely the point estimates to equal the true ones. In the estimation process, the random error is not avoidable. One way to account for is to compute p-values for a range of possible parameter values (including the null). The range of values, for which the p-value exceeds a specified alpha level (typically 0.05) is called confidence interval. An interval estimation procedure will, in 95% of repetitions (identical studies in all respects except for random error), produce limits that contain the true parameters. It is argued that the question if the pair of limits produced from a study contains the true parameter could not be answered by the ordinary (frequentist) theory of confidence intervals 1 . Frequentist approaches derive estimates by using probabilities of data (either p-values or likelihoods) as measures of compatibility between data and hypotheses, or as measures of the relative support that data provide hypotheses. Another approach, the Bayesian, uses data to improve existing (prior) estimates in light of new data. Proper use of any approach requires careful interpretation of statistics 1 , 2 .
The goal in any data analysis is to extract from raw information the accurate estimation. One of the most important and common question concerning if there is statistical relationship between a response variable (Y) and explanatory variables (Xi). An option to answer this question is to employ regression analysis in order to model its relationship. There are various types of regression analysis. The type of the regression model depends on the type of the distribution of Y; if it is continuous and approximately normal we use linear regression model; if dichotomous we use logistic regression; if Poisson or multinomial we use log-linear analysis; if time-to-event data in the presence of censored cases (survival-type) we use Cox regression as a method for modeling. By modeling we try to predict the outcome (Y) based on values of a set of predictor variables (Xi). These methods allow us to assess the impact of multiple variables (covariates and factors) in the same model 3 , 4 .
In this article we focus in linear regression. Linear regression is the procedure that estimates the coefficients of the linear equation, involving one or more independent variables that best predict the value of the dependent variable which should be quantitative. Logistic regression is similar to a linear regression but is suited to models where the dependent variable is dichotomous. Logistic regression coefficients can be used to estimate odds ratios for each of the independent variables in the model.
Linear equation
In most statistical packages, a curve estimation procedure produces curve estimation regression statistics and related plots for many different models (linear, logarithmic, inverse, quadratic, cubic, power, S-curve, logistic, exponential etc.). It is essential to plot the data in order to determine which model to use for each depedent variable. If the variables appear to be related linearly, a simple linear regression model can be used but in the case that the variables are not linearly related, data transformation might help. If the transformation does not help then a more complicated model may be needed. It is strongly advised to view early a scatterplot of your data; if the plot resembles a mathematical function you recognize, fit the data to that type of model. For example, if the data resemble an exponential function, an exponential model is to be used. Alternatively, if it is not obvious which model best fits the data, an option is to try several models and select among them. It is strongly recommended to screen the data graphically (e.g. by a scatterplot) in order to determine how the independent and dependent variables are related (linearly, exponentially etc.) 4 – 6 .
The most appropriate model could be a straight line, a higher degree polynomial, a logarithmic or exponential. The strategies to find an appropriate model include the forward method in which we start by assuming the very simple model i.e. a straight line (Y = a + bX or Y = b 0 + b 1 X ). Then we find the best estimate of the assumed model. If this model does not fit the data satisfactory, then we assume a more complicated model e.g. a 2nd degree polynomial (Y=a+bX+cX 2 ) and so on. In a backward method we assume a complicated model e.g. a high degree polynomial, we fit the model and we try to simplify it. We might also use a model suggested by theory or experience. Often a straight line relationship fits the data satisfactory and this is the case of simple linear regression. The simplest case of linear regression analysis is that with one predictor variable 6 , 7 .
Linear regression equation
The purpose of regression is to predict Y on the basis of X or to describe how Y depends on X (regression line or curve)

The Xi (X 1 , X 2 , , X k ) is defined as "predictor", "explanatory" or "independent" variable, while Y is defined as "dependent", "response" or "outcome" variable.
Assuming a linear relation in population, mean of Y for given X equals α+βX i.e. the "population regression line".
If Y = a + bX is the estimated line, then the fitted
Ŷi = a + bXi is called the fitted (or predicted) value, and Yi Ŷi is called the residual.
The estimated regression line is determined in such way that (residuals) 2 to be the minimal i.e. the standard deviation of the residuals to be minimized (residuals are on average zero). This is called the "least squares" method. In the equation

b is the slope (the average increase of outcome per unit increase of predictor)
a is the intercept (often has no direct practical meaning)
A more detailed (higher precision of the estimates a and b) regression equation line can also be written as

Further inference about regression line could be made by the estimation of confidence interval (95%CI for the slope b). The calculation is based on the standard error of b:

so, 95% CI for β is b ± t0.975*se(b) [t-distr. with df = n-2]
and the test for H0: β=0, is t = b / se(b) [p-value derived from t-distr. with df = n-2].
If the p value lies above 0.05 then the null hypothesis is not rejected which means that a straight line model in X does not help predicting Y. There is the possibility that the straight line model holds (slope = 0) or there is a curved relation with zero linear component. On the other hand, if the null hypothesis is rejected either the straight line model holds or in a curved relationship the straight line model helps, but is not the best model. Of course there is the possibility for a type II or type I error in the first and second option, respectively. The standard deviation of residual (σ res ) is estimated by

The standard deviation of residual (σ res ) characterizes the variability around the regression line i.e. the smaller the σ res , the better the fit. It has a number of degrees of freedom. This is the number to divide by in order to have an unbiased estimate of the variance. In this case df = n-2, because two parameters, α and β, are estimated 7 .
Multiple linear regression analysis
As an example in a sample of 50 individuals we measured: Y = toluene personal exposure concentration (a widespread aromatic hydrocarbon); X1 = hours spent outdoors; X2 = wind speed (m/sec); X3 = toluene home levels. Y is the continuous response variable ("dependent") while X1, X2, , Xp as the predictor variables ("independent") [7]. Usually the questions of interest are how to predict Y on the basis of the X's and what is the "independent" influence of wind speed, i.e. corrected for home levels and other related variables? These questions can in principle be answered by multiple linear regression analysis.
In the multiple linear regression model, Y has normal distribution with mean

The model parameters β 0 + β 1 + +β ρ and σ must be estimated from data.
β 0 = intercept
β 1 β ρ = regression coefficients
σ = σ res = residual standard deviation
Interpretation of regression coefficients
In the equation Y = β 0 + β 1 1 + +βρXρ
β 1 equals the mean increase in Y per unit increase in Xi , while other Xi's are kept fixed. In other words βi is influence of Xi corrected (adjusted) for the other X's. The estimation method follows the least squares criterion.
If b 0 , b 1 , , bρ are the estimates of β 0 , β 1 , , βρ then the "fitted" value of Y is

In our example, the statistical packages give the following estimates or regression coefficients (bi) and standard errors (se) for toluene personal exposure levels.

Then the regression equation for toluene personal exposure levels would be:

The estimated coefficient for time spent outdoors (0.582) means that the estimated mean increase in toluene personal levels is 0.582 g/m 3 if time spent outdoors increases 1 hour, while home levels and wind speed remain constant. More precisely one could say that individuals differing one hour in the time that spent outdoors, but having the same values on the other predictors, will have a mean difference in toluene xposure levels equal to 0.582 µg/m 3 8 .
Be aware that this interpretation does not imply any causal relation.
Confidence interval (CI) and test for regression coefficients
95% CI for i is given by bi ± t0.975*se(bi) for df= n-1-p (df: degrees of freedom)
In our example that means that the 95% CI for the coefficient of time spent outdoors is 95%CI: - 0.19 to 0.49

As in example if we test the H0: β humidity = 0 and find P = 0.40, which is not significant, we assumed that the association between between toluene personal exposure and humidity could be explained by the correlation between humididty and wind speed 8 .
In order to estimate the standard deviation of the residual (Y Yfit), i.e. the estimated standard deviation of a given set of variable values in a population sample, we have to estimate σ

The number of degrees of freedom is df = n (p + 1), since p + 1 parameters are estimated.
The ANOVA table gives the total variability in Y which can be partitioned in a part due to regression and a part due to residual variation:

With degrees of freedom (n 1) = p + (n p 1)
In statistical packages the ANOVA table in which the partition is given usually has the following format [6]:

SS: "sums of squares"; df: Degrees of freedom; MS: "mean squares" (SS/dfs); F: F statistics (see below)
As a measure of the strength of the linear relation one can use R. R is called the multiple correlation coefficient between Y, predictors (X1, Xp ) and Yfit and R square is the proportion of total variation explained by regression (R 2 =SSreg / SStot).
Test on overall or reduced model

In our example Tpers = β 0 + β 1 time outdoors + β 2 Thome +β 3 wind speed + residual
The null hypothesis (H 0 ) is that there is no regression overall i.e. β 1 = β 2 =+βρ = 0
The test is based on the proportion of the SS explained by the regression relative to the residual SS. The test statistic (F= MSreg / MSres) has F-distribution with df1 = p and df2 = n p 1 (F- distribution table). In our example F= 5.49 (P<0.01)
If now we want to test the hypothesis Ho: β 1 = β 2 = β 5 = 0 (k = 3)
In general k of p regression coefficients are set to zero under H0. The model that is valid if H 0 =0 is true is called the "reduced model". The Idea is to compare the explained variability of the model at hand with that of the reduced model.
The test statistic (F):

follows a F-distribution with df 1 = k and df 2 = n p 1.
If one or two variables are left out and we calculate SS reg (the statistical package does) and we find that the test statistic for F lies between 0.05 < P < 0.10, that means that there is some evidence, although not strong, that these variables together, independently of the others, contribute to the prediction of the outcome.
Assumptions
If a linear model is used, the following assumptions should be met. For each value of the independent variable, the distribution of the dependent variable must be normal. The variance of the distribution of the dependent variable should be constant for all values of the independent variable. The relationship between the dependent variable and the independent variables should be linear, and all observations should be independent. So the assumptions are: independence; linearity; normality; homoscedasticity. In other words the residuals of a good model should be normally and randomly distributed i.e. the unknown does not depend on X ("homoscedasticity") 2 , 4 , 6 , 9 .
Checking for violations of model assumptions
To check model assumptions we used residual analysis. There are several kinds of residuals most commonly used are the standardized residuals (ZRESID) and the studentized residuals (SRESID) [6]. If the model is correct, the residuals should have a normal distribution with mean zero and constant sd (i.e. not depending on X). In order to check this we can plot residuals against X. If the variation alters with increasing X, then there is violation of homoscedasticity. We can also use the Durbin-Watson test for serial correlation of the residuals and casewise diagnostics for the cases meeting the selection criterion (outliers above n standard deviations). The residuals are (zero mean) independent, normally distributed with constant standard deviation (homogeneity of variances) 4 , 6 .
To discover deviations form linearity and homogeneity of variables we can plot residuals against each predictor or against predicted values. Alternatively by using the PARTIAL plot we can assess linearity of a predictor variable. The partial plot for a predictor X 1 is a plot of residuals of Y regressed on other Xs and against residuals of Xi regressed on other X's. The plot should be linear. To check the normality of residuals we can use an histogram (with normal curve) or a normal probability plot 6 , 7 .
The goodness-of-fit of the model is assessed by studying the behavior of the residuals, looking for "special observations / individuals" like outliers, observations with high "leverage" and influential points. Observations deserving extra attention are outliers i.e. observations with unusually large residual; high leverage points: unusual x - pattern, i.e. outliers in predictor space; influential points: individuals with high influence on estimate or standard error of one or more β's. An observation could be all three. It is recommended to inspect individuals with large residual, for outliers; to use distances for high leverage points i.e. measures to identify cases with unusual combinations of values for the independent variables and cases that may have a large impact on the regression model. For influential points use influence statistics i.e. the change in the regression coefficients (DfBeta(s)) and predicted values (DfFit) that results from the exclusion of a particular case. Overall measure for influence on all β's jointly is "Cook's distance" (COOK). Analogously for standard errors overall measure is COVRATIO 6 .
Deviations from model assumptions
We can use some tips to correct some deviation from model assumptions. In case of curvilinearity in one or more plots we could add quadratic term(s). In case of non homogeneity of residual sd, we can try some transformation: log Y if Sres is proportional to predicted Y; square root of Y if Y distribution is Poisson-like; 1/Y if Sres 2 is proportional to predicted Y; Y 2 if Sres 2 decreases with Y. If linearity and homogeneity hold then non-normality does not matter if the sample size is big enough (n≥50- 100). If linearity but not homogeneity hold then estimates of β's are correct, but not the standard errors. They can be corrected by computing the "robust" se's (sandwich, Huber's estimate) 4 , 6 , 9 .
Selection methods for Linear Regression modeling
There are various selection methods for linear regression modeling in order to specify how independent variables are entered into the analysis. By using different methods, a variety of regression models from the same set of variables could be constructed. Forward variable selection enters the variables in the block one at a time based on entry criteria. Backward variable elimination enters all of the variables in the block in a single step and then removes them one at a time based on removal criteria. Stepwise variable entry and removal examines the variables in the block at each step for entry or removal. All variables must pass the tolerance criterion to be entered in the equation, regardless of the entry method specified. A variable is not entered if it would cause the tolerance of another variable already in the model to drop below the tolerance criterion 6 . In a model fitting the variables entered and removed from the model and various goodness-of-fit statistics are displayed such as R2, R squared change, standard error of the estimate, and an analysis-of-variance table.
Relative issues
Binary logistic regression models can be fitted using either the logistic regression procedure or the multinomial logistic regression procedure. An important theoretical distinction is that the logistic regression procedure produces all statistics and tests using data at the individual cases while the multinomial logistic regression procedure internally aggregates cases to form subpopulations with identical covariate patterns for the predictors based on these subpopulations. If all predictors are categorical or any continuous predictors take on only a limited number of values the mutinomial procedure is preferred. As previously mentioned, use the Scatterplot procedure to screen data for multicollinearity. As with other forms of regression, multicollinearity among the predictors can lead to biased estimates and inflated standard errors. If all of your predictor variables are categorical, you can also use the loglinear procedure.
In order to explore correlation between variables, Pearson or Spearman correlation for a pair of variables r (Xi, Xj) is commonly used. For each pair of variables (Xi, Xj) Pearson's correlation coefficient (r) can be computed. Pearsons r (Xi; Xj) is a measure of linear association between two (ideally normally distributed) variables. R 2 is the proportion of total variation of the one explained by the other (R 2 = b * Sx/Sy), identical with regression. Each correlation coefficient gives measure for association between two variables without taking other variables into account. But there are several useful correlation concepts involving more variables. The partial correlation coefficient between Xi and Xj, adjusted for other X's e.g. r (X1; X2 / X3). The partial correlation coefficient can be viewed as an adjustment of the simple correlation taking into account the effect of a control variable: r(X ; Y / Z ) i.e. correlation between X and Y controlled for Z. The multiple correlation coefficient between one X and several other X's e.g. r (X1 ; X2 , X3 , X4) is a measure of association between one variable and several other variables r (Y ; X1, X2, , Xk). The multiple correlation coefficient between Y and X1, X2,, Xk is defined as the simple Pearson correlation coefficient r (Y ; Yfit) between Y and its fitted value in the regression model: Y = β0 + β1X1+ βkXk + residual. The square of r (Y; X1, , Xk ) is interpreted as the proportion of variability in Y that can be explained by X1, , Xk. The null hypothesis [H 0 : ρ ( : X1, , Xk) = 0] is tested with the F-test for overall regression as it is in the multivariate regression model (see above) 6 , 7 . The multiple-partial correlation coefficient between one X and several other X`s adjusted for some other X's e.g. r (X1 ; X2 , X3 , X4 / X5 , X6 ). The multiple partial correlation coefficient equal the relative increase in % explained variability in Y by adding X1,, Xk to a model already containing Z1, , Zρ as predictors 6 , 7 .
Other interesting cases of multiple linear regression analysis include: the comparison of two group means. If for example we wish to answer the question if mean HEIGHT differs between men and women? In the simple linear regression model:

Testing β1 = 0 is equivalent with testing
HEIGHT MEN sub> = HEIGHT WOMEN by means of Student's t-test
The linear regression model assumes a normal distribution of HEIGHT in both groups, with equal . This is exactly the model of the two-sample t-test. In the case of comparison of several group means, we wish to answer the question if mean HEIGHT differ between different SES classes?
SES: 1 (low); 2 (middle) and 3 (high) (socioeconomic status)
We can use the following linear regression model:

Then β 1 and β 2 are interpreted as:
β 1 = difference in mean HEIGHT between low and high class
β 2 = difference in mean HEIGHT between middle and high class
Testing β 1 = β 2 = 0 is equivalent with the one-way ANalysis Of VAriance F-test . The statistical model in both cases is in fact the same 4 , 6 , 7 , 9 .
Analysis of covariance (ANCOVA)
If we wish to compare a continuous variable Y (e.g. HEIGHT) between groups (e.g. men and women) corrected (adjusted or controlled) for one or more covariables X (confounders) (e.g. X = age or weight) then the question is formulated: Are means of HEIGHT of men and women different, if men and women of equal weight are compared? Be aware that this question is different from that if there is a difference between the means of HEIGHT for men and women? And the answers can be quite different! The difference between men and women could be opposite, larger or smaller than the crude if corrected. In order to estimate the corrected difference the following multiple regression model is used:

where Y: response variable (for example HEIGHT); Z: grouping variable (for example Z = 0 for men and Z = 1 for women); X: covariable (confounder) (for example weight).
So, for men the regression line is y = β 0 + β 2 and for women is y = (β 0 + β 1 ) + β 2 .
This model assumes that regression lines are parallel. Therefore β 1 is the vertical difference, and can be interpreted as the: for X corrected difference between the mean response Y of the groups. If the regression lines are not parallel, then difference in mean Y depends on value of X. This is called "interaction" or "effect modification" .
A more complicated model, in which interaction is admitted, is:

regression line men: y = β 0 + β 2
regression line women: y = (β 0 + β 1 )+ (β 2 + β 3 )X
The hypothesis of the absence of "effect modification" is tested by H 0 : 3 = 0
As an example, we are interested to answer what is - the corrected for body weight - difference in HEIGHT between men and women in a population sample?
We check the model with interaction:

By testing β 3 =0, a p-value much larger than 0.05 was calculated. We assume therefore that there is no interaction i.e. regression lines are parallel. Further Analysis of Covariance for ≥ 3 groups could be used if we ask the difference in mean HEIGHT between people with different level of education (primary, medium, high), corrected for body weight. In a model where the three lines may be not parallel we have to check for interaction (effect modification) 7. Testing the hypothesis that coefficient of interactions terms equal 0, it is reasonable to assume a model without interaction. Testing the hypothesis H 0 : β 1 = β 2 = 0, i.e. no differences between education level when corrected for weight, gives the result of fitting the model, for which the P-values for Z 1 and Z 2 depend on your choice of the reference group. The purposes of ANCOVA are to correct for confounding and increase of precision of an estimated difference.
In a general ANCOVA model as:

where Y the response variable; k groups (dummy variables Z 1 , Z 2 , , Z k-1 ) and X 1 , , X p confounders
there is a straightforward extension to arbitrary number of groups and covariables.
Coding categorical predictors in regression
One always has to figure out which way of coding categorical factors is used, in order to be able to interpret the parameter estimates. In "reference cell" coding, one of the categories plays the role of the reference category ("reference cell"), while the other categories are indicated by dummy variables. The β's corresponding to the dummies that are interpreted as the difference of corresponding category with the reference category. In "difference with overall mean" coding in the model of the previous example: [Y = β 0 + β 1 1 + β 2 2 ++ residual], the β 0 is interpreted as the overall mean of the three levels of education while β 1 and β 2 are interpreted as the deviation of mean of primary and medium from overall mean, respectively. The deviation of the mean of high level from overall mean is given by (- β 1 - β 2 ). In "cell means" coding in the previous model (without intercept): [Y = β 0 + β 1 1 + β 2 2 + β 3 3 + residual], β 1 is the mean of primary, β 2 the middle and β 3 of the high level education 6 , 7 , 9 .
Conclusions
It is apparent to anyone who reads the medical literature today that some knowledge of biostatistics and epidemiology is a necessity. The goal in any data analysis is to extract from raw information the accurate estimation. But before any testing or estimation, a careful data editing, is essential to review for errors, followed by data summarization. One of the most important and common question is if there is statistical relationship between a response variable (Y) and explanatory variables (Xi). An option to answer this question is to employ regression analysis. There are various types of regression analysis. All these methods allow us to assess the impact of multiple variables on the response variable.
Quantitative Research Methods for Political Science, Public Policy and Public Administration: 4th Edition With Applications in R
11 introduction to multiple regression.
In the chapters in Part 3 of this book, we will introduce and develop multiple ordinary least squares regression – that is, linear regression models using two or more independent (or explanatory) variables to predict a dependent variable. Most users simply refer to it as “multiple regression”. 20 This chapter will provide the background in matrix algebra that is necessary to understand both the logic of, and notation commonly used for, multiple regression. As we go, we will apply the matrix form of regression in examples using R to provide a basic understanding of how multiple regression works. Chapter 12 will focus on the key assumptions about the concepts and data that are necessary for OLS regression to provide unbiased and efficient estimates of the relationships of interest, and it will address the key virtue of multiple regressions – the application of “statistical controls” in modeling relationships through the estimation of partial regression coefficients. Chapter 13 will turn to the process and set of choices involved in specifying and estimating multiple regression models, and to some of the automated approaches to model building you’d best avoid (and why). Chapter 13 turns to some more complex uses of multiple regression, such as the use and interpretation of “dummy” (dichotomous) independent variables, and modeling interactions in the effects of the independent variables. Chapter 14 concludes this part of the book with the application of diagnostic evaluations to regression model residuals, which will allow you to assess whether key modeling assumptions have been met and – if not – what the implications are for your model results. By the time you have mastered the chapters in this section, you will be well primed for understanding and using multiple regression analysis.
11.1 Matrix Algebra and Multiple Regression
Matrix algebra is widely used for the derivation of multiple regression because it permits a compact, intuitive depiction of regression analysis. For example, an estimated multiple regression model in scalar notion is expressed as: \(Y = A + BX_1 + BX_2 + BX_3 + E\) . Using matrix notation, the same equation can be expressed in a more compact and (believe it or not!) intuitive form: \(y = Xb + e\) .
In addition, matrix notation is flexible in that it can handle any number of independent variables. Operations performed on the model matrix \(X\) , are performed on all independent variables simultaneously. Lastly, you will see that matrix expression is widely used in statistical presentations of the results of OLS analysis. For all these reasons, then, we begin with the development of multiple regression in matrix form.
11.2 The Basics of Matrix Algebra
A matrix is a rectangular array of numbers with rows and columns. As noted, operations performed on matrices are performed on all elements of a matrix simultaneously. In this section we provide the basic understanding of matrix algebra that is necessary to make sense of the expression of multiple regression in matrix form.
11.2.1 Matrix Basics
The individual numbers in a matrix are referred to as “elements”. The elements of a matrix can be identified by their location in a row and column, denoted as \(A_{r,c}\) . In the following example, \(m\) will refer to the matrix row and \(n\) will refer to the column.
\(A_{m,n} = \begin{bmatrix} a_{1,1} & a_{1,2} & \cdots & a_{1,n} \\ a_{2,1} & a_{2,2} & \cdots & a_{2,n} \\ \vdots & \vdots & \ddots & \vdots \\ a_{m,1} & a_{m,2} & \cdots & a_{m,n} \end{bmatrix}\)
Therefore, in the following matrix;
\(A = \begin{bmatrix} 10 & 5 & 8 \\ -12 & 1 & 0 \end{bmatrix}\)
element \(a_{2,3} = 0\) and \(a_{1,2} = 5\) .
11.2.2 Vectors
A vector is a matrix with single column or row. Here are some examples:
\(A = \begin{bmatrix} 6 \\ -1 \\ 8 \\ 11 \end{bmatrix}\)
\(A = \begin{bmatrix} 1 & 2 & 8 & 7 \\ \end{bmatrix}\)
11.2.3 Matrix Operations
There are several “operations” that can be performed with and on matrices. Most of the these can be computed with R , so we will use R examples as we go along. As always, you will understand the operations better if you work the problems in R as we go. There is no need to load a data set this time – we will enter all the data we need in the examples.
11.2.4 Transpose
Transposing, or taking the “prime” of a matrix, switches the rows and columns. 21 The matrix
Once transposed is:
\(A' = \begin{bmatrix} 10 & -12 \\ 5 & 1 \\ 8 & 0 \end{bmatrix}\)
Note that the operation “hinges” on the element in the upper right-hand corner of \(A\) , \(A_{1,1}\) , so the first column of \(A\) becomes the first row on \(A'\) . To transpose a matrix in R , create a matrix object then simply use the t command.
11.2.5 Adding Matrices
To add matrices together, they must have the same dimensions , meaning that the matrices must have the same number of rows and columns. Then, you simply add each element to its counterpart by row and column. For example:
\(A = \begin{bmatrix} 4 & -3 \\ 2 & 0 \end{bmatrix} + B = \begin{bmatrix} 8 & 1 \\ 4 & -5 \end{bmatrix} = A+B = \begin{bmatrix} 4+8 & -3+1 \\ 2+4 & 0+(-5) \end{bmatrix} = \begin{bmatrix} 12 & -2 \\ 6 & -5 \end{bmatrix}\)
To add matrices together in R , simply create two matrix objects and add them together.
See – how easy is that? No need to be afraid of a little matrix algebra!
11.2.6 Multiplication of Matrices
To multiply matrices they must be conformable , which means the number of columns in the first matrix must match the number of rows in the second matrix.
Then, multiply column elements by the row elements, as shown here:
\(A = \begin{bmatrix} 2 & 5 \\ 1 & 0 \\ 6 & -2 \end{bmatrix} * B = \begin{bmatrix} 4 & 2 & 1 \\ 5 & 7 & 2 \end{bmatrix} = A X B = \\ \begin{bmatrix} (2 X 4)+(5 X 5) & (2 X 2)+(5 X 7) & (2 X 1)+(5 X 2) \\ (1 X 4)+(0 X 5) & (1 X 2)+(0 X 7) & (1 X 1)+(0 X 2) \\ (6 X 4)+(-2 X 5) & (6 X 2)+(-2 X 7) & (6 X 1)+(-2 X 2) \end{bmatrix} = \begin{bmatrix} 33 & 39 & 12 \\ 4 & 2 & 1 \\ 14 & -2 & 2 \end{bmatrix}\)
To multiply matrices in R , create two matrix objects and multiply them using the \%*\% command.
11.2.7 Identity Matrices
The identity matrix is a square matrix with 1’s on the diagonal and 0’s elsewhere. For a 4 x 4 matrix, it looks like this:
\(I = \begin{bmatrix} 1 & 0 & 0 & 0 \\ 0 & 1 & 0 & 0 \\ 0 & 0 & 1 & 0 \\ 0 & 0 & 0 & 1 \end{bmatrix}\)
It acts like a 1 in algebra; a matrix ( \(A\) ) times the identity matrix ( \(I\) ) is \(A\) . This can be demonstrated in R .
Note that, if you want to square a column matrix (that is, multiply it by itself), you can simply take the transpose of the column (thereby making it a row matrix) and multiply them. The square of column matrix \(A\) is \(A'A\) .
11.2.8 Matrix Inversion
The matrix inversion operation is a bit like dividing any number by itself in algebra. An inverse of the \(A\) matrix is denoted \(A^{-1}\) . Any matrix multiplied by its inverse is equal to the identity matrix:
For example,
\(A = \begin{bmatrix} 1 & -1 \\ -1 & -1 \end{bmatrix} \text{and } A^{-1} = \begin{bmatrix} 0.5 & -0.5 \\ -0.5 & 0.5 \end{bmatrix} \text{therefore } A*A^{-1} = \begin{bmatrix} 1 & 0 \\ 0 & 1 \end{bmatrix}\)
However, matrix inversion is only applicable to a square (i.e., number of rows equals number of columns) matrix; only a square matrix can have an inverse.
Finding the Inverse of a Matrix
To find the inverse of a matrix, the values that will produce the identity matrix, create a second matrix of variables and solve for \(I\) .
\(A = \begin{bmatrix} 3 & 1 \\ 2 & 4 \end{bmatrix} X \begin{bmatrix} a & b \\ c & d \end{bmatrix} = \begin{bmatrix} 3a+b & 3c+d \\ 2a+4b & 2c+4d \end{bmatrix} = \begin{bmatrix} 1 & 0 \\ 0 & 1 \end{bmatrix}\)
Set \(3a+b=1\) and \(2a+4b=0\) and solve for \(a\) and \(b\) . In this case \(a = \frac{2}{5}\) and \(b = -\frac{1}{5}\) . Likewise, set \(3c+d=0\) and \(2c+4d=1\) ; solving for \(c\) and \(d\) produces \(c=-\frac{1}{10}\) and \(d=\frac{3}{10}\) . Therefore,
\(A^{-1} = \begin{bmatrix} \frac{2}{5} & -\frac{1}{10} \\ -\frac{1}{5} & \frac{3}{10} \end{bmatrix}\)
Finding the inverse matrix can also be done in R using the solve command.
OK – now we have all the pieces we need to apply matrix algebra to multiple regression.
11.3 OLS Regression in Matrix Form
As was the case with simple regression, we want to minimize the sum of the squared errors, \(e\) . In matrix notation, the OLS model is \(y=Xb+e\) , where \(e = y-Xb\) . The sum of the squared \(e\) is:
Therefore, we want to find the \(b\) that minimizes this function:
To do this we take the derivative of \(e'e\) w.r.t \(b\) and set it equal to \(0\) .
Then to remove the \(-2\) ’s, we multiply each side by \(-1/2\) . This leaves us with:
To solve for \(b\) we multiply both sides by the inverse of \(X'X, (X'X)^{-1}\) . Note that for matrices this is equivalent to dividing each side by \(X'X\) . Therefore:
The \(X'X\) matrix is square, and therefore invertible (i.e., the inverse exists). However, the \(X'X\) matrix can be non-invertible (i.e., singular) if \(n < k\) —the number of \(k\) independent variables exceeds the \(n\) -size—or if one or more of the independent variables is perfectly correlated with another independent variable. This is termed perfect multicollinearity and will be discussed in more detail in Chapter 14. Also note that the \(X'X\) matrix contains the basis for all the necessary means, variances, and covariances among the \(X\) ’s.
Regression in Matrix Form
- \(y=n*1\) column vector of observations of the DV, \(Y\)
- \(\hat{y}=n*1\) column vector of predicted \(Y\) values
- \(X=n*k\) matrix of observations of the IVs; first column \(1\) s
- \(b=k*1\) column vector of regression coefficients; first row is \(A\)
- \(e=n*1\) column vector of \(n\) residual values
Using the following steps, we will use R to calculate \(b\) , a vector of regression coefficients; \(\hat y\) , a vector of predicted \(y\) values; and \(e\) , a vector of residuals.
We want to fit the model \(y = Xb+e\) to the following matrices:
\[ y = \begin{bmatrix} 6 \\ 11 \\ 4 \\ 3 \\ 5 \\ 9 \\ 10 \end{bmatrix}\quad X = \begin{bmatrix} 1 & 4 & 5 & 4 \\ 1 & 7 & 2 & 3 \\ 1 & 2 & 6 & 4 \\ 1 & 1 & 9 & 6 \\ 1 & 3 & 4 & 5 \\ 1 & 7 & 3 & 4 \\ 1 & 8 & 2 & 5 \end{bmatrix} \]
Create two objects, the \(y\) matrix and the \(X\) matrix.
Calculate \(b\) : \(b = (X'X)^{-1}X'y\) .
We can calculate this in R in just a few steps. First, we transpose \(X\) to get \(X'\) .
Then we multiply \(X\) by \(X'\) ; ( \(X'X\) ).
Next, we find the inverse of \(X'X\) ; \(X'X^{-1}\)
Then, we multiply \(X'X^{-1}\) by \(X'\) .
Finally, to obtain the \(b\) vector we multiply \(X'X^{-1}X'\) by \(y\) .
We can use the lm function in R to check and see whether our “by hand” matrix approach gets the same result as does the “canned” multiple regression routine:
Calculate \(\hat y\) : \(\hat y=Xb\) .
To calculate the \(\hat y\) vector in R , simply multiply X and b .
Calculate \(e\) .
To calculate \(e\) , the vector of residuals, simply subtract the vector \(y\) from the vector \(\hat y\) .

11.4 Summary
Whew! Now, using matrix algebra and calculus, you have derived the squared-error minimizing formula for multiple regression. Not only that, you can use the matrix form, in R , to calculate the estimated slope and intercept coefficients, predict \(Y\) , and even calculate the regression residuals. We’re on our way to true Geekdome!
Next stop: the key assumptions necessary for OLS to provide the best, unbiased, linear estimates (BLUE) and the basis for statistical controls using multiple independent variables in regression models.
It is useful to keep in mind the difference between “multiple regression” and “multivariate regression”. The latter predicts 2 or more dependent variables using an independent variable. ↩
The use of “prime” in matrix algebra should not be confused with the use of ``prime" in the expression of a derivative, as in \(X'\) . ↩
The Multiple Regression Analysis
- First Online: 04 September 2024
Cite this chapter

- Franz Kronthaler 2
In the previous chapter, we saw that usually one independent variable is not sufficient to adequately describe the dependent variable. Normally, several factors have an influence on the dependent variable. Hence, typically we need multiple regression analysis, also known as multivariate regression analysis, to describe an issue.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Author information
Authors and affiliations.
Hochschule Graubünden FHGR, Chur, Switzerland
Franz Kronthaler
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Franz Kronthaler .
Rights and permissions
Reprints and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer-Verlag GmbH, DE, part of Springer Nature
About this chapter
Kronthaler, F. (2024). The Multiple Regression Analysis. In: Statistics Applied with the R Commander. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-662-69107-6_20
Download citation
DOI : https://doi.org/10.1007/978-3-662-69107-6_20
Published : 04 September 2024
Publisher Name : Springer, Berlin, Heidelberg
Print ISBN : 978-3-662-69106-9
Online ISBN : 978-3-662-69107-6
eBook Packages : Mathematics and Statistics Mathematics and Statistics (R0)
Share this chapter
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research

Regression Analysis
Regression analysis is a quantitative research method which is used when the study involves modelling and analysing several variables, where the relationship includes a dependent variable and one or more independent variables. In simple terms, regression analysis is a quantitative method used to test the nature of relationships between a dependent variable and one or more independent variables.
The basic form of regression models includes unknown parameters (β), independent variables (X), and the dependent variable (Y).
Regression model, basically, specifies the relation of dependent variable (Y) to a function combination of independent variables (X) and unknown parameters (β)
Y ≈ f (X, β)
Regression equation can be used to predict the values of ‘y’, if the value of ‘x’ is given, and both ‘y’ and ‘x’ are the two sets of measures of a sample size of ‘n’. The formulae for regression equation would be
Do not be intimidated by visual complexity of correlation and regression formulae above. You don’t have to apply the formula manually, and correlation and regression analyses can be run with the application of popular analytical software such as Microsoft Excel, Microsoft Access, SPSS and others.
Linear regression analysis is based on the following set of assumptions:
1. Assumption of linearity . There is a linear relationship between dependent and independent variables.
2. Assumption of homoscedasticity . Data values for dependent and independent variables have equal variances.
3. Assumption of absence of collinearity or multicollinearity . There is no correlation between two or more independent variables.
4. Assumption of normal distribution . The data for the independent variables and dependent variable are normally distributed
My e-book, The Ultimate Guide to Writing a Dissertation in Business Studies: a step by step assistance offers practical assistance to complete a dissertation with minimum or no stress. The e-book covers all stages of writing a dissertation starting from the selection to the research area to submitting the completed version of the work within the deadline. John Dudovskiy

Multiple Regression Analysis using SPSS Statistics
Introduction.
Multiple regression is an extension of simple linear regression. It is used when we want to predict the value of a variable based on the value of two or more other variables. The variable we want to predict is called the dependent variable (or sometimes, the outcome, target or criterion variable). The variables we are using to predict the value of the dependent variable are called the independent variables (or sometimes, the predictor, explanatory or regressor variables).
For example, you could use multiple regression to understand whether exam performance can be predicted based on revision time, test anxiety, lecture attendance and gender. Alternately, you could use multiple regression to understand whether daily cigarette consumption can be predicted based on smoking duration, age when started smoking, smoker type, income and gender.
Multiple regression also allows you to determine the overall fit (variance explained) of the model and the relative contribution of each of the predictors to the total variance explained. For example, you might want to know how much of the variation in exam performance can be explained by revision time, test anxiety, lecture attendance and gender "as a whole", but also the "relative contribution" of each independent variable in explaining the variance.
This "quick start" guide shows you how to carry out multiple regression using SPSS Statistics, as well as interpret and report the results from this test. However, before we introduce you to this procedure, you need to understand the different assumptions that your data must meet in order for multiple regression to give you a valid result. We discuss these assumptions next.
SPSS Statistics
Assumptions.
When you choose to analyse your data using multiple regression, part of the process involves checking to make sure that the data you want to analyse can actually be analysed using multiple regression. You need to do this because it is only appropriate to use multiple regression if your data "passes" eight assumptions that are required for multiple regression to give you a valid result. In practice, checking for these eight assumptions just adds a little bit more time to your analysis, requiring you to click a few more buttons in SPSS Statistics when performing your analysis, as well as think a little bit more about your data, but it is not a difficult task.
Before we introduce you to these eight assumptions, do not be surprised if, when analysing your own data using SPSS Statistics, one or more of these assumptions is violated (i.e., not met). This is not uncommon when working with real-world data rather than textbook examples, which often only show you how to carry out multiple regression when everything goes well! However, don’t worry. Even when your data fails certain assumptions, there is often a solution to overcome this. First, let's take a look at these eight assumptions:
- Assumption #1: Your dependent variable should be measured on a continuous scale (i.e., it is either an interval or ratio variable). Examples of variables that meet this criterion include revision time (measured in hours), intelligence (measured using IQ score), exam performance (measured from 0 to 100), weight (measured in kg), and so forth. You can learn more about interval and ratio variables in our article: Types of Variable . If your dependent variable was measured on an ordinal scale, you will need to carry out ordinal regression rather than multiple regression. Examples of ordinal variables include Likert items (e.g., a 7-point scale from "strongly agree" through to "strongly disagree"), amongst other ways of ranking categories (e.g., a 3-point scale explaining how much a customer liked a product, ranging from "Not very much" to "Yes, a lot").
- Assumption #2: You have two or more independent variables , which can be either continuous (i.e., an interval or ratio variable) or categorical (i.e., an ordinal or nominal variable). For examples of continuous and ordinal variables , see the bullet above. Examples of nominal variables include gender (e.g., 2 groups: male and female), ethnicity (e.g., 3 groups: Caucasian, African American and Hispanic), physical activity level (e.g., 4 groups: sedentary, low, moderate and high), profession (e.g., 5 groups: surgeon, doctor, nurse, dentist, therapist), and so forth. Again, you can learn more about variables in our article: Types of Variable . If one of your independent variables is dichotomous and considered a moderating variable, you might need to run a Dichotomous moderator analysis .
- Assumption #3: You should have independence of observations (i.e., independence of residuals ), which you can easily check using the Durbin-Watson statistic, which is a simple test to run using SPSS Statistics. We explain how to interpret the result of the Durbin-Watson statistic, as well as showing you the SPSS Statistics procedure required, in our enhanced multiple regression guide.
- Assumption #4: There needs to be a linear relationship between (a) the dependent variable and each of your independent variables, and (b) the dependent variable and the independent variables collectively . Whilst there are a number of ways to check for these linear relationships, we suggest creating scatterplots and partial regression plots using SPSS Statistics, and then visually inspecting these scatterplots and partial regression plots to check for linearity. If the relationship displayed in your scatterplots and partial regression plots are not linear, you will have to either run a non-linear regression analysis or "transform" your data, which you can do using SPSS Statistics. In our enhanced multiple regression guide, we show you how to: (a) create scatterplots and partial regression plots to check for linearity when carrying out multiple regression using SPSS Statistics; (b) interpret different scatterplot and partial regression plot results; and (c) transform your data using SPSS Statistics if you do not have linear relationships between your variables.
- Assumption #5: Your data needs to show homoscedasticity , which is where the variances along the line of best fit remain similar as you move along the line. We explain more about what this means and how to assess the homoscedasticity of your data in our enhanced multiple regression guide. When you analyse your own data, you will need to plot the studentized residuals against the unstandardized predicted values. In our enhanced multiple regression guide, we explain: (a) how to test for homoscedasticity using SPSS Statistics; (b) some of the things you will need to consider when interpreting your data; and (c) possible ways to continue with your analysis if your data fails to meet this assumption.
- Assumption #6: Your data must not show multicollinearity , which occurs when you have two or more independent variables that are highly correlated with each other. This leads to problems with understanding which independent variable contributes to the variance explained in the dependent variable, as well as technical issues in calculating a multiple regression model. Therefore, in our enhanced multiple regression guide, we show you: (a) how to use SPSS Statistics to detect for multicollinearity through an inspection of correlation coefficients and Tolerance/VIF values; and (b) how to interpret these correlation coefficients and Tolerance/VIF values so that you can determine whether your data meets or violates this assumption.
- Assumption #7: There should be no significant outliers , high leverage points or highly influential points . Outliers, leverage and influential points are different terms used to represent observations in your data set that are in some way unusual when you wish to perform a multiple regression analysis. These different classifications of unusual points reflect the different impact they have on the regression line. An observation can be classified as more than one type of unusual point. However, all these points can have a very negative effect on the regression equation that is used to predict the value of the dependent variable based on the independent variables. This can change the output that SPSS Statistics produces and reduce the predictive accuracy of your results as well as the statistical significance. Fortunately, when using SPSS Statistics to run multiple regression on your data, you can detect possible outliers, high leverage points and highly influential points. In our enhanced multiple regression guide, we: (a) show you how to detect outliers using "casewise diagnostics" and "studentized deleted residuals", which you can do using SPSS Statistics, and discuss some of the options you have in order to deal with outliers; (b) check for leverage points using SPSS Statistics and discuss what you should do if you have any; and (c) check for influential points in SPSS Statistics using a measure of influence known as Cook's Distance, before presenting some practical approaches in SPSS Statistics to deal with any influential points you might have.
- Assumption #8: Finally, you need to check that the residuals (errors) are approximately normally distributed (we explain these terms in our enhanced multiple regression guide). Two common methods to check this assumption include using: (a) a histogram (with a superimposed normal curve) and a Normal P-P Plot; or (b) a Normal Q-Q Plot of the studentized residuals. Again, in our enhanced multiple regression guide, we: (a) show you how to check this assumption using SPSS Statistics, whether you use a histogram (with superimposed normal curve) and Normal P-P Plot, or Normal Q-Q Plot; (b) explain how to interpret these diagrams; and (c) provide a possible solution if your data fails to meet this assumption.
You can check assumptions #3, #4, #5, #6, #7 and #8 using SPSS Statistics. Assumptions #1 and #2 should be checked first, before moving onto assumptions #3, #4, #5, #6, #7 and #8. Just remember that if you do not run the statistical tests on these assumptions correctly, the results you get when running multiple regression might not be valid. This is why we dedicate a number of sections of our enhanced multiple regression guide to help you get this right. You can find out about our enhanced content as a whole on our Features: Overview page, or more specifically, learn how we help with testing assumptions on our Features: Assumptions page.
In the section, Procedure , we illustrate the SPSS Statistics procedure to perform a multiple regression assuming that no assumptions have been violated. First, we introduce the example that is used in this guide.
A health researcher wants to be able to predict "VO 2 max", an indicator of fitness and health. Normally, to perform this procedure requires expensive laboratory equipment and necessitates that an individual exercise to their maximum (i.e., until they can longer continue exercising due to physical exhaustion). This can put off those individuals who are not very active/fit and those individuals who might be at higher risk of ill health (e.g., older unfit subjects). For these reasons, it has been desirable to find a way of predicting an individual's VO 2 max based on attributes that can be measured more easily and cheaply. To this end, a researcher recruited 100 participants to perform a maximum VO 2 max test, but also recorded their "age", "weight", "heart rate" and "gender". Heart rate is the average of the last 5 minutes of a 20 minute, much easier, lower workload cycling test. The researcher's goal is to be able to predict VO 2 max based on these four attributes: age, weight, heart rate and gender.
Setup in SPSS Statistics
In SPSS Statistics, we created six variables: (1) VO 2 max , which is the maximal aerobic capacity; (2) age , which is the participant's age; (3) weight , which is the participant's weight (technically, it is their 'mass'); (4) heart_rate , which is the participant's heart rate; (5) gender , which is the participant's gender; and (6) caseno , which is the case number. The caseno variable is used to make it easy for you to eliminate cases (e.g., "significant outliers", "high leverage points" and "highly influential points") that you have identified when checking for assumptions. In our enhanced multiple regression guide, we show you how to correctly enter data in SPSS Statistics to run a multiple regression when you are also checking for assumptions. You can learn about our enhanced data setup content on our Features: Data Setup page. Alternately, see our generic, "quick start" guide: Entering Data in SPSS Statistics .
Test Procedure in SPSS Statistics
The seven steps below show you how to analyse your data using multiple regression in SPSS Statistics when none of the eight assumptions in the previous section, Assumptions , have been violated. At the end of these seven steps, we show you how to interpret the results from your multiple regression. If you are looking for help to make sure your data meets assumptions #3, #4, #5, #6, #7 and #8, which are required when using multiple regression and can be tested using SPSS Statistics, you can learn more in our enhanced guide (see our Features: Overview page to learn more).
Note: The procedure that follows is identical for SPSS Statistics versions 18 to 28 , as well as the subscription version of SPSS Statistics, with version 28 and the subscription version being the latest versions of SPSS Statistics. However, in version 27 and the subscription version , SPSS Statistics introduced a new look to their interface called " SPSS Light ", replacing the previous look for versions 26 and earlier versions , which was called " SPSS Standard ". Therefore, if you have SPSS Statistics versions 27 or 28 (or the subscription version of SPSS Statistics), the images that follow will be light grey rather than blue. However, the procedure is identical .

Published with written permission from SPSS Statistics, IBM Corporation.
Note: Don't worry that you're selecting A nalyze > R egression > L inear... on the main menu or that the dialogue boxes in the steps that follow have the title, Linear Regression . You have not made a mistake. You are in the correct place to carry out the multiple regression procedure. This is just the title that SPSS Statistics gives, even when running a multiple regression procedure.

Interpreting and Reporting the Output of Multiple Regression Analysis
SPSS Statistics will generate quite a few tables of output for a multiple regression analysis. In this section, we show you only the three main tables required to understand your results from the multiple regression procedure, assuming that no assumptions have been violated. A complete explanation of the output you have to interpret when checking your data for the eight assumptions required to carry out multiple regression is provided in our enhanced guide. This includes relevant scatterplots and partial regression plots, histogram (with superimposed normal curve), Normal P-P Plot and Normal Q-Q Plot, correlation coefficients and Tolerance/VIF values, casewise diagnostics and studentized deleted residuals.
However, in this "quick start" guide, we focus only on the three main tables you need to understand your multiple regression results, assuming that your data has already met the eight assumptions required for multiple regression to give you a valid result:
Determining how well the model fits
The first table of interest is the Model Summary table. This table provides the R , R 2 , adjusted R 2 , and the standard error of the estimate, which can be used to determine how well a regression model fits the data:

The " R " column represents the value of R , the multiple correlation coefficient . R can be considered to be one measure of the quality of the prediction of the dependent variable; in this case, VO 2 max . A value of 0.760, in this example, indicates a good level of prediction. The " R Square " column represents the R 2 value (also called the coefficient of determination), which is the proportion of variance in the dependent variable that can be explained by the independent variables (technically, it is the proportion of variation accounted for by the regression model above and beyond the mean model). You can see from our value of 0.577 that our independent variables explain 57.7% of the variability of our dependent variable, VO 2 max . However, you also need to be able to interpret " Adjusted R Square " ( adj. R 2 ) to accurately report your data. We explain the reasons for this, as well as the output, in our enhanced multiple regression guide.
Statistical significance
The F -ratio in the ANOVA table (see below) tests whether the overall regression model is a good fit for the data. The table shows that the independent variables statistically significantly predict the dependent variable, F (4, 95) = 32.393, p < .0005 (i.e., the regression model is a good fit of the data).

Estimated model coefficients
The general form of the equation to predict VO 2 max from age , weight , heart_rate , gender , is:
predicted VO 2 max = 87.83 – (0.165 x age ) – (0.385 x weight ) – (0.118 x heart_rate ) + (13.208 x gender )
This is obtained from the Coefficients table, as shown below:

Unstandardized coefficients indicate how much the dependent variable varies with an independent variable when all other independent variables are held constant. Consider the effect of age in this example. The unstandardized coefficient, B 1 , for age is equal to -0.165 (see Coefficients table). This means that for each one year increase in age, there is a decrease in VO 2 max of 0.165 ml/min/kg.
Statistical significance of the independent variables
You can test for the statistical significance of each of the independent variables. This tests whether the unstandardized (or standardized) coefficients are equal to 0 (zero) in the population. If p < .05, you can conclude that the coefficients are statistically significantly different to 0 (zero). The t -value and corresponding p -value are located in the " t " and " Sig. " columns, respectively, as highlighted below:

You can see from the " Sig. " column that all independent variable coefficients are statistically significantly different from 0 (zero). Although the intercept, B 0 , is tested for statistical significance, this is rarely an important or interesting finding.
Putting it all together
You could write up the results as follows:
A multiple regression was run to predict VO 2 max from gender, age, weight and heart rate. These variables statistically significantly predicted VO 2 max, F (4, 95) = 32.393, p < .0005, R 2 = .577. All four variables added statistically significantly to the prediction, p < .05.
If you are unsure how to interpret regression equations or how to use them to make predictions, we discuss this in our enhanced multiple regression guide. We also show you how to write up the results from your assumptions tests and multiple regression output if you need to report this in a dissertation/thesis, assignment or research report. We do this using the Harvard and APA styles. You can learn more about our enhanced content on our Features: Overview page.

Quantitative Research Methods
- Introduction
- Descriptive and Inferential Statistics
- Hypothesis Testing
- Regression and Correlation
- Time Series
- Meta-Analysis
- Mixed Methods
- Additional Resources
- Get Research Help

Correlation is the relationship or association between two variables. There are multiple ways to measure correlation, but the most common is Pearson's correlation coefficient (r), which tells you the strength of the linear relationship between two variables. The value of r has a range of -1 to 1 (0 indicates no relationship). Values of r closer to -1 or 1 indicate a stronger relationship and values closer to 0 indicate a weaker relationship. Because Pearson's coefficient only picks up on linear relationships, and there are many other ways for variables to be associated, it's always best to plot your variables on a scatter plot, so that you can visually inspect them for other types of correlation.
- Correlation Penn State University tutorial
- Correlation and Causation Australian Bureau of Statistics Article
Spurious Relationships
It's important to remember that correlation does not always indicate causation. Two variables can be correlated without either variable causing the other. For instance, ice cream sales and drownings might be correlated, but that doesn't mean that ice cream causes drownings—instead, both ice cream sales and drownings increase when the weather is hot. Relationships like this are called spurious correlations.
- Spuriousness Harvard Business Review article.
- New Evidence for Theory of The Stork A satirical article demonstrating the dangers of confusing correlation with causation.

Regression is a statistical method for estimating the relationship between two or more variables. In theory, regression can be used to predict the value of one variable (the dependent variable) from the value of one or more other variables (the independent variable/s or predictor/s). There are many different types of regression, depending on the number of variables and the properties of the data that one is working with, and each makes assumptions about the relationship between the variables. (For instance, most types of regression assume that the variables have a linear relationship.) Therefore, it is important to understand the assumptions underlying the type of regression that you use and how to properly interpret its results. Because regression will always output a relationship, whether or not the variables are truly causally associated, it is also important to carefully select your predictor variables.
- A Refresher on Regression Analysis Harvard Business Review article.
- Introductory Business Statistics - Regression
Simple Linear Regression
Simple linear regression estimates a linear relationship between one dependent variable and one independent variable.
- Simple Linear Regression Tutorial Penn State University Tutorial
- Statistics 101: Linear Regression, The Very Basics YouTube video from Brandon Foltz.
Multiple Linear Regression
Multiple linear regression estimates a linear relationship between one dependent variable and two or more independent variables.
- Multiple Linear Regression Tutorial Penn State University Tutorial
- Multiple Regression Basics NYU course materials.
- Statistics 101: Multiple Linear Regression, The Very Basics YouTube video from Brandon Foltz.
If you do a subject search for Regression Analysis you'll see that the library has over 200 books about regression. Select books are listed below. Also, note that econometrics texts will often include regression analysis and other related methods.
Search for ebooks using Quicksearch . Use keywords to search for e-books about Regression .
- << Previous: Hypothesis Testing
- Next: ANOVA >>
- Last Updated: Aug 16, 2024 1:12 PM
- URL: https://guides.library.duq.edu/quant-methods
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 06 September 2024
An integrated analysis of multiple datasets reveals novel gene signatures in human granulosa cells
- Xhulio Dhori 1 , 2 ,
- Silvia Gioiosa ORCID: orcid.org/0000-0003-1302-8320 1 na1 &
- Stefania Gonfloni ORCID: orcid.org/0000-0002-9392-4258 2 na1
Scientific Data volume 11 , Article number: 972 ( 2024 ) Cite this article
Metrics details
- Cell signalling
- Reproductive biology
- RNA sequencing
Granulosa cells (GCs) play crucial roles in oocyte maturation. Through gap junctions and extracellular vesicles, they mediate the exchange of molecules such as microRNAs and messenger RNAs. Different ovarian cell types exhibit unique gene expression profiles, reflecting their specialized functions and stages. By combining RNA-seq data from various cell types forming the follicle, we aimed at capturing a wide range of expression patterns, offering insights into the functional diversity and complexity of the transcriptome regulation across GCs. Herein, we performed an integrated bioinformatics analysis of RNA sequencing datasets present in public databases, with a unique and standardized workflow., By combining the data from different studies, we successfully increased the robustness and reliability of our findings and discovered novel genes, miRNAs, and signaling pathways associated with GCs function and oocyte maturation. Moreover, our results provide a valuable resource for further wet-lab research on GCs biology and their impact on oocyte development and competence.
Introduction
Ovarian follicles are the functional units of the ovary. Within the follicle, the bidirectional signaling between somatic cells and the oocyte changes continuously over time to synchronize follicle development and oocyte maturation. Integrated signaling cascades coordinate oocyte maturation, regulating GC proliferation and theca cell differentiation 1 . Yet, the function and the roles of distinct types of intercellular communications remain poorly clarified. Follicle maturation is a complex dynamic process, tightly controlled through autocrine and paracrine regulatory factors produced principally by theca, granulosa cells and oocytes. In addition, it is controlled by hormones (from the hypothalamic-pituitary-ovarian axis, including pituitary –derived FSH, LH) and steroids secreted by the ovary. GCs play an essential role during oocyte maturation, providing an optimal microenvironment for follicle development 2 , 3 , 4 . Two major cell types of GCs, named mural and cumulus, surround respectively the follicular antrum and the oocyte. Mural granulosa cells (MGCs), have an endocrine function and promote follicle growth, whereas cumulus cells (CCs) support oocyte maturation and provide essential metabolic and signaling functions 5 , 6 . Although MGCs and CCs are closely related granulosa cell types, differences in their function and gene expression profile 5 suggest that these significant changes, in gene expression, coordinate the follicle maturation 7 .
As the oocyte competence is acquired only through bidirectional communication with GCs 2 , the closely surrounding CCs constitute an attractive biological material for all the molecular analyses finalized to infer on the developmental abilities of the oocyte. This implies that CCs transcriptomic profiling may serve to identify biomarkers for predicting oocyte quality 8 . Several studies have described gene expression of CCs (matured either in vivo or in vitro) in cumulus-oocyte complex (COC) 9 or derived from pre-ovulatory follicles exposed to human chorionic gonadotrophin (hCG) 5 , 10 , 11 .
For example, Yerushalmi et al . (PRJNA216966) 12 have reported transcriptional differences between CCs derived from mid-antral follicles (compact cumulus cells) and expanded CCs from metaphase 2 (M2) COC, identifying 1746 Differentially Expressed Genes (DEGs). In addition, they found 89 Differentially Expressed (DE) long noncoding RNAs, a few of them encoded within introns of genes required for GC function. For what concerns transcriptional gene expression profiles within COC, Li et al . (PRJNA649934) 13 performed RNA-seq experiments to investigate the transcription gene profile of CCs and Oocytes derived from the same Polycystic Ovary syndrome (PCOS) patient or from age-matched controls. In PCOS oocyte, they found an altered expression of some genes involved in Microtubule-based processes 13 . Overall, global gene expression in CCs and in oocytes from PCOS patients showed imbalance in other essential signalling pathways underlying Mitochondrial Function and Oxidative Stress.
MicroRNAs (miRNAs) modulate gene expression post-transcriptionally, yet their expression and function within the GC populations remain poorly investigated. Recent reports described and studied the miRNA profile of MGCs and CCs isolated from human pre-ovulatory follicles from healthy women undergoing ovum pick up for in vitro fertilization (IVF). Velthut-Meikas et al . (PRJNA200696) 14 found 90 miRNAs differently expressed between CGCs and MGCs, [2 among them were of intronic origin]. In MGCs, some of the DE miRNAs-targets specifically act on extracellular matrix remodeling, Wnt/beta-catenin and Neurotrophin signaling pathways.
A more recent study by Andrei et al . (PRJNA417973) 15 reported the miRNA expression profile of human primary MGCs and CCs, in vitro cultured. In this study, the authors found 53 miRNAs differentially expressed between MGCs and CCs, whose targets are enriched in Ubiquitin-like Protein Conjugation genes. Interestingly, the two studies showed a limited overlap in terms of DE miRNAs, mostly because of differences in data analysis strategy.
Nevertheless, the overview of the published studies describing CCs’ transcriptome has provided an extremely limited consensus of “signatures”.
Conversely, by combining RNA-seq data from various cell types, we can capture a wide range of expression patterns, offering insights into the functional diversity and complexity of the transcriptome expression and regulation across the GCs, Secondly, comparative transcriptional profiling of CCs derived from immature or mature COC would help identifying genes that are expressed in the final stages of follicle maturation.
Through a survey of publicly available transcriptomics datasets, we re-analysed the raw RNA-seq data from different “sources”: two GC subpopulations, two CCs samples derived from mature and immature COCs, and from CCs and the oocyte. The original studies laid a solid foundation, but we saw an opportunity to delve deeper, asking new questions that bring out the latent possibilities within these datasets. Thereby, we also focused on Differential Alternative Splicing (DAS) analysis of CCs transcripts, to get insights into alternative splicing events occurring during follicle maturation. By revisiting and reinterpreting the data, we aimed to uncover additional layers of biological insight, providing a richer and more nuanced understanding of the biological landscapes under study. This approach not only maximizes the value of the existing data but also opens new avenues for discovery and innovation, such as the prediction of some biomarkers for measuring the oocyte competence and reproductive potential.
We selected a cohort of samples according to inclusion criteria detailed under the Methods section. The final core dataset consists of 51 samples, (30 miRNAs samples and 21 mRNAs) for a total of 98,82 GB of input data (Table 1 , Supplementary Table 1 ). Raw data from the selected BioProjects were downloaded from Sequence Read Archive (SRA) 16 and individually reanalyzed applying the same bioinformatics pipeline depicted in Fig. 1 .

Workflow diagram of the RNA-seq analysis. Overview of the data analysis pipeline. The pipeline begins with the acquisition of raw reads in FASTQ format. These reads are then assessed for quality using the FastQC program. Terminal low-quality bases and adapter sequences are subsequently trimmed off. Mapped files in BAM format are then generated from the processed reads and raw read counts are quantified. The pipeline proceeds with differential gene expression analysis. The resulting differentially expressed genes (DEGs) are analysed for Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) enrichment. Starting from BAM input files, the pipeline also includes a parallel branch analysis for identifying Differential Alternative Spliced Genes (DAS), enriched by a splicing events distribution analysis and a GO- and KEGG- enrichment analysis for DAS. Finally, miRNA analysis is conducted as part of the small-RNA seq process. This comprehensive pipeline provides a robust approach to RNA-Seq data analysis, from raw reads to functional insights..
In the present study, three RNA-sequencing datasets from GCs were reanalyzed to identify DEGs (Differentially Expressed Genes) and DAS (Differentially Alternatively Spliced Genes). In addition, to evaluate the miRNAs profile of granulosa cells we re-analyzed and integrated two further Bioprojects treating small-RNASeq data (Table 1 ).
Actin-binding proteins (ABPs) play crucial roles in granulosa cell function and oocyte maturation
For cumulus cells, three different bioprojects were analyzed for Gene Expression Analysis (Table 1 ).
Firstly, in PRJNA216966, we identified 3093 DEGs, nearly double the number of DEGs previously reported 12 , with 1662 genes up-regulated and 1431 down-regulated in Germinal Vesicle stage CC compared to M2-expanded CCs (Supplementaty Fig. 1a,b ).
Secondly, in PRJNA200696, 2242 DEGs were identified, consisting of 1201 up-regulated and 1041 down-regulated genes in CCs compared to MGCs (Supplementaty Fig. 2a,b ).
Lastly, in PRJNA649934, the differential expression analysis detected 7253 DEGs, with 3609 genes up-regulated and 3644 down-regulated in Oocytes compared to CCs (Supplementaty Fig. 3a,b ).
As a first step, an intra -Bioproject functional enrichment analysis was performed. This approach ensured that the biological context of each specific dataset was considered independently, minimizing the potential confounding effects of combining disparate conditions. Supplementary figures illustrate these analyses (Fig S1 1c, 1d, S2 2c, 2d, S3c, 3d).
Beyond the individual project re-analysis, we performed a comparative analysis to understand the distribution of DEGs across different cell types, including cumulus cells, oocytes, and mural cells. This comparative approach was carefully designed to highlight both the unique and the shared pathways/biological processes among the different cell types. This approach helped us identify critical pathways and mechanisms that might be overlooked if we only focused on one direction of gene regulation.
Therefore, an inter -bioprojects comparison of functional profiles was executed to gain insights into the biological roles of DEGs emerging from the three BioProjects. By including both up- and down-regulated genes in the gene set enrichment analysis, we could better understand how these regulatory changes impact the biological functions and communication networks essential for follicle development. This analysis allowed us both to find a functional consensus and, at the same time, to highlight any differential functional module specific for each data set (Fig. 2a ).

Analysis and comparison of enriched GO terms for DEGs. ( a ) Dotplot shows enriched GO terms over the 3 BioProjects, highlighting the first twelve most significantly over-represented GO terms in biological processes. The x-axis reports the Bioproject name and the n. of DEGs involved in those GO processes reported in parenthesis. The y-axis shows significantly enriched GO terms. The size of each dot indicates the number of genes associated with that GO term in the respective cluster (gene count). The color scale represents the adjusted p-value, with darker colors indicating higher statistical significance. Only GO terms with an adjusted p-value < 0.05 are shown. ( b ) Upset plot of the intersection of DEGs across BioProjects. The horizontal bar graph on the left shows the total number of DEGs for each BioProject. The upper black bar graphs show the number of DEGs for each overlapping combination and black connected dots indicate which BioProjects are involved in each intersection. Plot shows the number of shared DEGs among 3 Bioprojects (n = 236, herein defined as “core set”), among at least two BioProjects (n = 390, 521 and 1229) and BioProject-specific (n = 1095, 1236 and 5267). ( c ) PPI network of the core set of 236 shared DEGs. The network nodes represent proteins. Colored nodes highlighted in red (n = 160) are proteins known to be involved in “Alternative splicing” with a great statistical significance (p-adj < 0.05).
The functional consensus analysis highlighted “Actin binding” as the GO Term shared among all comparisons, while the other GO Terms are more BioProject-specific or denote CC-specific GO Terms. Of note, CCs collected at two different developmental stages (GV-M2 CCs) are placed along a middle ground between pre-ovulatory CCs vs MCGs and expanded CCs vs Oocyte (Fig. 2a ).
To further elucidate the gene expression patterns within different cell types, we examined the distribution of up-regulated genes (Fig. S4 ), which advised our subsequent focus on CCs. Therefore, we performed a functional enrichment analysis solely on genes that were up regulated in CCs (taken at different stages of follicle maturation) and results are reported in Fig. S5 . This targeted approach ensured the functional analysis to be cumulus-specific, without contamination from markers of mural GCs or oocytes. Results of this more focused analysis highlighted roughly the same enriched categories, like “actin binding” and different terms related to cell-cell communication and metabolic activity, which are known to be critical in CC function and oocyte maturation. Additionally, we identified both shared and unique pathways across different stages of cumulus cell development.
PPI analysis reveals a coreset of 236 DEGs potentially involved in alternative splicing processes
We were also interested in using a complementary approach to GO-enrichment analysis for finding a consensus list of proteins which could be key interactors and regulators of different developmental stages of CCs. As shown in the upset plot in Fig. 2b , there is a coreset of 236 DEGs shared by the three Bioprojects (Fig. 2b , first bar in the upset plot; the whole list of overlappings is supplied 17 .
To this end, we used STRING database 18 which focuses on experimentally validated and literature-derived protein-protein interactions. It provides insights into how proteins interact and form functional complexes, highlighting biological processes and pathways that might not be as apparent in GO analysis. Intriguingly, by applying a PPI (Protein-Protein Interaction) network analysis on the core set, the “Alternative Splicing” pattern resulted enormously enriched with an FDR of 9.96e-05 (Fig. 2c , red nodes) thus suggesting that, besides individual peculiarities of each Bioproject, these 236 DEGs could have strong consequences on Alternative splicing variations in GCs.
Unraveling the impact of alternative splicing events on transcription regulators
Previous analyses of RNA-seq data in GCs subpopulations have largely focused on gene expression rather than alternative splicing. Nevertheless, the results obtained in our PPI analysis strongly encouraged us to investigate the extent to which alternative splicing contributes to transcriptomic variation between each data subsets.
In CCs from the GV cumulus-oocyte-complexes (COCs) and from the M2 expanded COCs (PRJNA216966), Differential Alternative Splicing analysis identified 1402 DAS (Supplementary Table 2 ), including 104 transcription factors (TFs).
In pre-ovulatory follicles, (CCs vs MCGs) (PRJNA200696), we identified 378 DAS events (Supplementary Table 2 ), involving 45 TFs. In stimulated vs unstimulated CCs (PRJNA649934), we detected 924 DAS events (Supplementary Table 2 ), 72 of which are TFs.
Regarding the splicing event distribution of each Bioproject (Fig. 3a ), we found more similarity between PRJNA200696 and PRJNA216966, probably because both bioprojects share as source the same granulosa cells type. Particularly, Intron Retention (IR) and Multiple Exon Skipping (MES) events seem to have a great role in AS events occurring in GCs. Conversely, in PRJNA649934 we found a greater enrichment in terms of Alternative Last Exons (ALE).

Analysis of splicing event distribution and comparison of enriched GO terms and KEGGs for DAS. ( a ) Splicing distribution events over the DAS detected in the three mRNA bioprojects showing cumulative percentage in the histogram. The categories are: IR (Intron Retention); ALE/AFE (Alternative Last/First Exon); MES (Multiple Exon skipping); MXE (Mutually Exclusive Exons); alt3′ (alternative 3′ exon); alt5′ (alternative 5′ exon). ( b ) A dotplot that shows the common enriched GO terms for DAS in the three bioprojects. The x-axis represents different CC clusters, while the y-axis shows significantly enriched GO terms. The size of each dot indicates the number of genes associated with that GO term in the respective cluster (gene count). The color scale represents the adjusted p-value, with darker colors indicating higher statistical significance. Only GO terms with an adjusted p-value < 0.05 are shown. ( c ) The cnet-plot shows common GO Terms and highlights the identity of the DAS that contributes to the enrichment of that specific category. ( d ) The cnet-plot shows common KEGG Pathways and highlights the identity of the DAS that contributes to the enrichment of that specific category.
As consequence, a cluster comparison of Gene Ontology and KEGG enrichment analysis of the DAS lists shows greater similarities between PRJNA200696 and PRJNA216966, while PRJNA649934 has specific enriched GO terms (Supplementary Table 3 ). In pre-ovulatory follicles, CCs versus MCGs, and in stimulated versus unstimulated CCs, alternative splicing affects genes involved in “retinoic acid receptor binding” (GO:0042974), “nuclear hormone receptor binding” (GO:0035257) (Fig. 3b,c ) and “Ubiquitin mediated proteolysis” (hsa04120) Kegg pathways (Fig. 3d ).
On the other hand, “Cadherin binding” GO term results strongly enriched for DAS in Cumulus Cells from GV cumulus-oocyte-complex (COC) and from M2 expanded COC and in stimulated versus unstimulated CCs (Fig. 3b,c ).
Specific pathways uniquely enriched in PRJNA216966, are “Ubiquitin binding”, “DNA-binding transcription factor binding” and “RNA polymerase II-specific DNA-binding transcription factor binding” (Fig. 3b ). Therefore, given the high impact that this last category of DAS (n = 104) could have on transcription, we analyzed in greater detail the expression of downstream targets. By applying a GO enrichment analysis over the DEGs targeted by the 104 TF-DAS, we found some terms in common with the ontological DEGs analysis (GO:0001816 cytokine production, GO:0030155, regulation of cell adhesion) and intriguingly some unique terms like “female sex differentiation” (GO:0046660), “ovarian follicle development” (GO:0001541) and “reproductive structure development” (GO:0048608).
Given the impact on transcription highlighted in Fig. 3c , we wondered whether we could observe significant changes in terms of log2FC on the target genes of DAS Transcription Regulators. Although we have found some cases in which DAS Transcription Regulators seemed to have an effect over the cumulative gene expression of its target genes (Suppl. Figure 6a ), our analysis pointed out that Differential Splicing of TFs may not be the only event to determine effects on downstream transcription.
Nevertheless, to identify potentially interesting genes for future studies, we used the VOILA web-based application to focus our attention on the AS events affecting some TFs which resulted interesting from the Supplementary Fig. 6a . We observed that many DAS events fall in regions transcribing functional protein domains associated with epigenetic modifications, notably histone modification.
We performed an additional GO enrichment analysis on DEG targets of DAS Transcription Factors for each Bioproject.
Interestingly we found “Transcription coregulator activity” and “Transcription factor binding” largely enriched in PRJNA216966 in Fig. 3c . Alternative splicing is a fine-tuning regulatory mechanism that can vary significantly between similar cell types or different maturation stages. In fact, in the previously mentioned dataset comparing cumulus cells at GV (germinal vesicle) and M2 (metaphase II) stages, we found numerous GO terms related to key processes such as “ovarian follicle development,” “response to follicle-stimulating hormone,” and different terms related to cell differentiation (Fig. S6b ). These findings highlight the role of alternative splicing in regulating the genes involved in these critical biological processes and the importance of transcription regulators in orchestrating the complex events leading to ovulation. Enrichment results of the PRJNA200696 and PRJNA649934 are shown in Figure S6c,d . Among the DAS-TFs found in PRJNA216966, [CCs in two developmental stages, GV-M2], GATA4 (GATA Binding Protein 4), GATA6 (GATA Binding Protein 6), NR5A1 (nuclear receptor subfamily 5 group A member 1), NR5A2 (nuclear receptor subfamily 5 group A member 2), all these TFs play a crucial role in follicle maturation (Fig. 4 ).

Splicegraph analysis and dPSI of functional splicing events occurring in transcriptional regulators in PRJNA216966. Each plot provides a visual representation of a splicegraph along with the differential Percent Spliced In (dPSI) values for a specific splice junction. The plot emphasizes splice junctions exhibiting significant dPSI values, with annotations indicating their associated biological functions within the highlighted region. The splice graph is a directed acyclic graph representing alternative splicing events in a gene. It illustrates connections between exons and introns: exons are denoted by numbered nodes enclosed in boxes, while intron retention events are represented by smaller boxes lacking numerical labels. Raw read counts for each specific splicing event are displayed on both nodes and edges. At the top of each plot, the genomic coordinates of the splicing junctions are indicated. At the bottom of the plot, a violin plot is presented, illustrating the distribution of differential Percent Spliced In (dPSI) values associated with the highlighted junction. The width of the violin plot conveys the probability density of different dPSI values, offering insights into the variability of splicing patterns. ( a ) GATA6. ( b ) GATA4. ( c ) NR5A1. ( d ) NR5A2.
miRNA DE analysis reveals a core set of 12 DEMs which may influence cell cycle progression and nucleic acids metabolism
Both the bioprojects (PRJNA417973; PRJNA200699) have investigated the miRNA transcriptome landscape of CCs and MGCs. This allowed us to make an integrated complete comparison for searching either meta-signatures of differentially expressed miRNAs (DEMs) or common DEMs to use for GO and KEGG enrichment analysis.
In detail, the analysis of PRJNA417973 highlighted 46 DEMs, half of which (23) up-regulated and the other half down-regulated (Fig. 5a ); while in PRJNA200699 we found more DEMs in CCs compared to MCs: 82 DEMs (Fig. 5b ), 48 among them up-regulated and 34 down-regulated.

PRJNA200699 and PRJNA417973 small RNA-seq analysis. ( a ) Heatmap of differential expressed miRNAs in PRJNA200699. ( b ) Heatmap of differential expressed miRNAs in PRJNA417973. ( c ) Venn diagram illustrating miRNAs that have the same expression pattern between the two bioprojects (intersection, herein indicated as DEM-core set, n = 12) and Bioproject-specific DEMs. ( d ) Common enriched GO terms over the target DEGs of DEM-core set. ( e ) PRJNA200699 network shows DEM-core set and targets which are differentially expressed. Spheres represent miRNAs colored by log2FC, from red up-regulation to blue down-regulation.
Intersecting the two lists of DEMs, we found 12 common DEMs which interestingly share the same pattern of expression, unveiling cell type specific “signatures”. In MGCs hsa-miR-30a-5p, hsa-miR-30a-3p, hsa-miR-142-5p, hsa-miR-148a-3p, hsa-miR-10b-5p, hsa-miR-144-5p and hsa-miR-146a-5p are up-regulated while hsa-miR-92a-1-5p, hsa-miR-145-5p, hsa-miR-149-5p, hsa-miR-129-5p and hsa-miR-196a-5p are all up-regulated in CCs.
Since the pattern of expression was extremely coherent in the two granulosa cell types, we exerted a GO enrichment analysis over the predicted target genes of the 12 shared DEMs to enlighten downstream pathways modulated by such miRNA core set within the two GC populations. The GO analysis highlighted common enriched GO-terms with statistical significance, first among them, “histone modification (GO:0016570)”, “regulation of mRNA metabolic process (GO:1903311)”, “regulation of DNA metabolic process (GO:0051052)”, “regulation of translation (GO:0006417)”, “response to decreased oxygen (GO:0036293)”, “mitotic cell cycle phase transition (GO:)”(Fig. 5d ).
Moreover, access to an independent gene expression dataset (PRJNA200696) that compares mural and cumulus cells provided a significant advantage in our analysis. This dataset enabled direct evaluation and validation of whether the pathways regulated by miRNA in cumulus and mural cells do exhibit corresponding changes at the mRNA level. Our analysis revealed a significant overlap in Gene Ontology (GO) terms between the targets of the identified miRNAs and the mRNA expression data from the independent dataset (Fig. S8 ) thus indicating that the pathways regulated by these miRNAs in cumulus and mural cells are also reflected at the mRNA level in the independent dataset.
To further elucidate the regulatory network of miRNAs and their target genes, we constructed a miRNA-mRNA interaction network based on our differential expression analyses results (Fig. 5e ). The resulting network was visualized using the Fruchterman-Reingold algorithm, a force-directed layout that positions nodes based on their connections. In this visualization, miRNA positioning reflects the number of edges (target connections), with more centrally located miRNAs typically having a higher number of target mRNAs. Notably, neighboring miRNAs in the network tend to share more common mRNA targets, suggesting potential co-regulatory relationships. This network approach allowed us to identify key miRNA regulators, based on their connectivity, and to discern patterns of miRNA-mRNA interactions that may be biologically relevant to our experimental conditions.
Our study investigates various cell types under their physiological conditions, with a particular focus on the interactions between mural/cumulus cells and the oocyte within the follicle. These cells communicate to coordinate follicle maturation and other essential processes. Consequently, we examined the entire follicle system, analyzing how genes are upregulated or downregulated within these cells, ultimately contributing to key biological processes involved in follicle maturation.
To identify specific signatures associated with each granulosa cell type, we reanalyzed raw data obtained from several selected publicly available bioprojects using a standardized transcriptomic computational workflow. We aimed to analyze different datasets to gain insights into the enrichment of differentially expressed genes between human MGCs and CCs from distinct stages [in the Germinal Vesicle stage cumulus cells (GV-CCs) compared to the M2 expanded cumulus cells (M2-CCs) and the oocyte compared to the CCs].
After obtaining the list of DEGs specific for each Bioproject, a biological theme comparison was used to compare functional profiles from different conditions. Our rationale for including both up- and down-regulated genes in the GO enrichment analysis was to capture a comprehensive view of the regulatory mechanisms at play which could be crucial for the maturation and function of the follicle. Up-regulated genes can indicate processes that are being promoted or activated, while down-regulated genes can indicate processes that are being repressed or modulated.However, many processes are more complex than that: up and down regulated genes can participate in the same process simultaneously. Together, they provide a holistic picture of the cellular environment and the interactions between different cell types within the follicle.
Results indicate that the functional consensus module “Actin binding” is the GO-Term shared among all comparisons. To address the concern about potential cross-contamination from other cell types, we also focused exclusively on up-regulated genes in cumulus cells and conducted a thorough functional enrichment analysis (Fig. S5 ). “Actin Binding” was correctly remarked in this analysis as well, thus ensuring that the functional relevance of our results can be accurately attributed to cumulus cells. Remarkably, actin filaments are involved in multiple cellular processes such as cytoskeleton organization, nuclear positioning, germinal vesicle breakdown, spindle migration, chromosome segregation and polar body extrusion in oocyte mammalian meiosis 19 .
ABPs modulate the polymerization and depolymerization of actin filaments, thereby controlling the structure and dynamics of the actin cytoskeleton, and the secretory and endocytic pathway 20 . This regulation is vital for maintaining the structural integrity and shape of granulosa cells and oocytes 21 . Furthermore, during folliculogenesis, the development of ovarian follicles, granulosa cells proliferate and differentiate. ABPs help in remodeling the actin cytoskeleton, which is necessary for the migration, proliferation, and differentiation of granulosa cells, thus supporting follicle development 22 .
ABPs are involved in the formation and maintenance of gap junctions, ensuring proper signaling between GCs and oocytes. At the same time, the secretion of signaling molecules ensures nutrient exchange, which is critical for oocyte growth and maturation 1 .
MGCs, when stimulated to differentiate through FSH and insulin-like growth factors 23 , 24 , are involved in the endocrine function necessary for supporting the development of the follicle.
Notably, our comparative analysis of DEGs from pre-ovulatory MGCs vs CCs (Supplementary Fig. 2c,d ) has highlighted the following GO terms: “extracellular matrix structural constituent”, “extracellular matrix binding”, “glycosaminoglycan binding” and “collagen binding” 17 . This comparison reveals a clear correlation between these DEGs and the physiological events underlying the follicle maturation.
The extracellular matrix (ECM) plays a critical role in mammalian ovarian function and oogenesis 25 , 26 , 27 , 28 , while the MGCs line the antrum, the CCs surround the oocyte, being regulated by oocyte secreted factors (OSFs) 29 . In the pre-ovulatory follicle maturation process, CCs produce hyaluronic acid that is deposited into the extracellular matrix and later stabilized by secreted proteins 30 , 31 . This newly formed ECM connects the oocyte and cumulus cells together. The growing oocyte then derives most of its substrates for energy metabolism and biosynthesis from the surrounding CCs 32 , 33 .
Gap junctions transmit metabolites, nutrients, and intracellular signalling molecules and all the OSFs that are required for the expansion of CCs 34 . In turn, CCs synchronize nuclear and cytoplasm maturation of the oocyte and regulate meiosis resumption by providing many factors and regulatory molecules to oocytes 34 . Cumulus expansion is an important aspect in the final steps of follicle maturation, which culminates in the formation of a mature cumulus-oocyte complex arrested at the metaphase of the second meiotic division (M2) and ready for ovulation and its subsequent fertilization 35 .
Among the key factors that affect the “oocyte competence” are the cytoplasmic synthesis/degradation of maternal mRNA together with an ordered distribution of organelles 36 , 37 , 38 , 39 . The outcome of both processes affects fertilization and embryonic development 1 , 40 , 41 , 42 , 43 , 44 . Abnormal mitochondrial rearrangement impairs the oocyte quality and maturation and chromosomal separation during meiosis 45 , 46 . Cytoplasmic oocyte development also includes maturation of cortical granules 47 , 48 (membranous organelles derived from Golgi complexes) and cytoskeleton (microtubules and microfilaments network 49 ) as well as of the endoplasmic reticulum (ER), the latter being responsible for the storage and release of free calcium, essential for the calcium reaction at fertilization 50 , 51 . Compelling evidence indicates that Actin, Microtubule and Organelle Dynamics as well as Golgi distribution are fundamental for oocyte maturation 52 , 53 . By comparing GV oocyte-vs-CCs and CCs GV-vs-M2, we found the following enriched GO terms “Microtubule binding”, “tubulin binding”, “catalytic activity, acting on DNA”. Conversely, GO terms like “histone binding”, and “transcription co-regulator activity” were enriched only when comparing GV_Oocyte versus CCs. The GO term “histone binding” points out that epigenetic modifications play important roles following meiotic maturation of mammalian oocytes 54 .
In our search for key regulators of different developmental stages in CCs, we identified a core set of 236 DEGs, shared by the three Bioprojects.
We are aware that much of the complexity within cells arises from functional and regulatory interactions among proteins, therefore we wanted to conduct a complementary approach to GO-term analysis, namely the Protein-Protein Interaction (PPI) analysis. Interestingly, PPI network analysis of the core set indicates that the “Alternative Splicing” pattern resulted in being significantly enriched. We investigated the extent to which alternative splicing contributes to transcriptomic differences within all data sets.
To this end, we used the STRING database which focuses on experimentally validated and literature-derived protein-protein interactions. It provides insights into how proteins interact and form functional complexes, highlighting biological processes and pathways that might not be as apparent in GO analysis. The enrichment of “alternative splicing” term in STRING might indicate that the proteins involved in this process are highly interconnected and functionally significant within the shared coreset of 236 DEGs because many proteins interact directly in this process.
On the other hand, the solely GO enrichment analysis was not able to capture this important feature, probably because while it provides a broad view of functional categories, it might not always capture specific processes if they are not the main factor distinguishing these clusters.
Because of this result, we were strongly encouraged to perform a Differential Alternative Splicing (DAS) analysis on these data, which is something that none of the authors of the three Bioprojects have investigated in their published papers.
Regarding the DAS analysis we first wanted to describe the overall splicing changes in the three datasets, categorizing them into 8 categories and reporting cumulative frequencies. We observed a similar splicing event distribution when comparing the two bioprojects (PRJNA200696 and PRJNA216966), both based on a common granulosa cells source. In contrast, DAS events, in PRJNA649934, reveal a greater enrichment in terms of Alternative Last Exons (ALE) and, at the same time, a lower percentage of Mutually Exclusive Exons and Intron Retention events (IR). This observation could be linked to the mechanisms by which maternal mRNAs are stored in oocytes and gradually consumed with initiation of meiotic resumption. Alternative 3′ UTR isoforms enable inclusion or exclusion of cis-regulatory elements either for RNA binding proteins or microRNAs that can influence transcript abundance, stability, subcellular localization, and translation efficiency 55 . A large amount of maternal mRNA exists in mature oocytes at the GV stage although as dormant transcripts until meiotic maturation 56 , 57 . Compelling evidence supports the existence of stringent mRNA stabilizing mechanisms within GV-Oocytes, while the selective degradation of maternal mRNA is required for the activation of zygotic genome 39 , 58 . Regulation of maternal mRNA translation and degradation occurs during oocyte maturation and such events are essential for the oocyte competence necessary to accomplish maternal zygotic transition 36 , 59 . In maturing oocytes, cytoplasmic polyadenylation of the 3′-UTR is linked to mRNA stability and mRNA translation 60 . As such, we analyzed DAS events involving transcription regulators, including NR5A1, NR5A2, GATA4 and GATA6, and found that they affect domains of such TFs associated with epigenetic modification (i.e. histone modification). We have used differentially alternative spliced transcription factors (DAS-TFs) and their targets, which are differentially expressed genes (DEGs) within each dataset, for GO enrichment analysis. Our analysis revealed that many of the enriched GO terms are directly related to follicle development. The analysis was conducted on each individual dataset. This approach was necessary because alternative splicing is a fine-tuning regulatory mechanism that can vary significantly between similar cell types or different maturation stages. In fact, in our dataset comparing cumulus cells at GV (germinal vesicle) and M2 (metaphase II) stages, we found numerous GO terms related to key processes such as “ovarian follicle development,” “response to follicle-stimulating hormone,” and different terms related to cell differentiation. These findings highlight the role of alternative splicing in regulating the genes involved in these critical biological processes and the importance of transcription regulators in orchestrating the complex events leading to ovulation. Among the DAS-TFs, comparing cumulus cells at two developmental stages (GV_M2), we found GATA4, GATA6, NR5A1, NR5A2 (see Fig. 4 ). Compelling evidence indicates that GATA4 and GATA6 play a crucial role in folliculogenesis 61 . Although they do not contribute equally to ovarian function, as assessed by experiments performed with conditional knockout mice. Experimental results indicate that GATA4 regulates directly or indirectly more genes than GATA6. In particular, the expression of FSHR (which is essential for the differentiation of GCs) decreases only in GATA4 knockout mice. However, the role of GATA factors in promoting the expression of genes for extracellular matrix organization, steroid metabolism and ovulation has been clearly demonstrated using microarrays analysis. NR5A1 and NR5A2 are master regulators of steroidogenesis 62 , 63 . Defects in steroid hormone production impair follicular development and fertility. Experiments performed with conditional knockout mice indicate that the lack of expression of NR5A1 in postnatal ovary causes infertility 64 . Interestingly, also the overexpression of NR5A1 affects female fertility and metabolism, confirming the importance of a fine-tuned control of NR5A1 for female reproductive and metabolic functions 62 . Despite the structure similarity, NR5A1 and NR5A2 exert different effects in human granulosa cells. It has been proposed that the fine-tuning of steroidogenesis in the ovarian follicles is achieved through BMP-15 signaling that induces NR5A -target genes expression while suppressing NR5A2-linked genes 63 .
Interestingly, “Cadherin binding” GO terms result in being strongly enriched for DAS in stimulated versus unstimulated CCs and in CCs from GV-M2 COC. As mentioned above, folliculogenesis strictly depends on the contact of the surrounding granulosa cells and the oocyte. Cadherins are a group of membrane proteins involved in cell adhesion. They assure adhesion between adjacent cells by the homotypic interactions of cells exposing similar sets of cadherins at their surfaces 65 . Cadherins contribute to tissue integrity 66 , regulating cell migration, cell differentiation and the control size of a specific cell population 67 . Cadherin-catenin complexes also act as receptors for signaling molecules. Compelling evidence indicates that E- and N–cadherin together with associated catenin take part in NF-kb-mediated signaling, RhoA GTPase signaling, and Hippo, Yap1, RTK and Hedgehog pathways 68 , 69 , 70 , 71 . Interestingly, the Wnt4/beta-catenin cascade is crucial for ovary development in mice and other vertebrates 72 . Although it can be assumed that cadherins take part in the follicle maturation, since they constitute the core of adherens junctions, little is known about the signaling pathways regulated by cadherins during oocyte development. Therefore, our findings suggest a new deeper investigation of Alternative Splicing events in Cadherin-signaling associated genes. Despite the indirect suggestions of enriched follicle development function from DEGs only GO enrichment analysis, DAS analysis reported a significant enrichment of the ovarian follicle development category (GO:0001541) 73 .
How do maternal mRNAs remain stable during completion of meiosis or in initial stages of embryo development and how dormant transcripts are activated and degraded later? Likely, RNA-binding proteins are accumulated in fully grown oocytes for stabilizing and degrading mRNAs together with other components required for regulating protein synthesis and degradation. Interestingly, we observed several DAS genes involved in mRNA surveillance pathways by comparing GV_Oocyte vs CCs (Fig. 3d ). We found genes encoding RNA binding proteins that are components of multi protein Exon Junction Complex (EJC) placed at the splice junction on mRNAs (MAGOHB, RNPS1, ACIN1) and involved in the nonsense-mediated decay (NMD) of mRNAs (MAGOHB, RNPS1). RNPS1 participates in mRNA 3′-end cleavage and mediated an increase in RNA abundance and translational efficiency. DAS genes encode proteins that bind with high affinity to nascent poly(A) tails and stimulate poly (A) addition (PABPN1, FIPL1) and are involved in nucleocytoplasmic trafficking. Furthermore, CSTF1 and CSTF3 (Cleavage Stimulation Factor Subunit 1 and 3 respectively) genes encode two of the three subunits forming the cleavage stimulation factors (CSTF), which are involved in polyadenylation and cleavage of 3′ end of mammalian pre-mRNAs. Among DAS genes, we also found GSPT1 (G1 to S phase transition 1) gene that is involved in regulation of translation termination and predicted to be part of translation release factor complex 74 .
Furthermore, we found regulatory components of phosphorylation-mediated signaling involved in the negative control of cell growth and division (Protein Phosphatase 2 Regulatory Subunit B gamma, PPP2R3C and Protein Phosphatase 2 Scaffold Subunit Abeta, PPP2R1B, both genes present alternative splice transcript variants).
Several DAS genes associated with Ubiquitin mediated proteolysis were shared by comparing both GV_Oocyte vs CCs and CCs GV vs M2 COC. The ubiquitination/deubiquitination process is important for degradation of proteins, transcriptional regulation, and cell cycle progression 75 which are also crucial for the oocyte maturation. APC (anaphase-promoting complex) initiates the metaphase to anaphase transition by promoting cyclin B and securin degradation 76 . Among DAS genes, we found several core subunits of APC, like ANAPC5, ANAPC7, ANAPC10, ANAPC13 (which are large E3 ubiquitin ligases), controlling cell progression by targeting cyclin B for 26 S proteasome-mediated degradation. Through comparison of DAS between CCs_vs M2_CCs we found ANAPC13, ANPC7 and ANAPC5. Interestingly, multiple transcript variants for ANAPC5 gene have been described, resulting in shorter isoforms due to downstream AUG sites. Compelling evidence supports that ubiquitin E3 ligases mediate specific protein degradation, crucial in the progress of both meiotic and mitotic cell cycle 77 . Cullin ring-finger ubiquitin ligase 4 is one of the E3 ligase members that play multiple functions both in oocyte survival and in the meiotic cell cycle progression 76 . In line with this, among DAS shared by both groups (GV_Oocyte vs CCs and CCs_vs M2_CCs), we found CUL4A, ELOC, TRIP12M MGRN1, CDC27. The latter (together with CDC16) is a component of the anaphase-promoting complex (APC) that catalyzes the formation of cyclin B-ubiquitin conjugate ending in the ubiquitin-mediated proteolysis of B-type cyclins. TRIP12 is an E3 ubiquitin ligase implicated in the degradation of p19ARF/ARF isoform of CDKN2A and in the DNA Damage Response by regulating UP7 stability and by doing so of p53. Interestingly, among DAS shared by comparing CCs GV_M2 and GV_Oocyte vs CCs, we found several members of E2 Ubiquitin-conjugating enzyme E2 family (UBE2D3, UBE3B, UBE2E2, UBE2N, UBE2G). UBE2E2 is involved in positive regulation of G1/S transition of mitotic cell cycle, UBE2N can interact with UBE2V1/UBE2V2, catalyzing the synthesis of non-canonical Lys 63-coupled poly-ubiquitin chains. This type of poly-ubiquitination does not lead to protein degradation while mediating transcriptional activation of target genes. UBE2G2 mediates endoplasmic reticulum-associated degradation (ERAD) 78 , such as the sterol-induced ubiquitination of 3-hydroxy-3-methylglytaryl CoA reductase 79 . Among DAS genes derived from GV oocyte-vs-CCs comparison we found several E3 ubiquitin ligases. FBXW11 (F-box and WD Repeat Domain Containing 11), a substrate component of a SCF (SKPI-CUL1-F-box protein) that mediates the ubiquitination of phosphorylated proteins, allowing transcriptional activation. BIRC2 (Baculoviral IAP Repeat Containing 2), an E3 ubiquitin ligase that modulates mitogenic kinase signaling, cell proliferation and apoptosis. In addition, BIRC2 can stimulate the transcriptional activity of E2F1 and can also function as an E3 ligase for the NEDD8 conjugation pathway of effector caspases. SYVN1 (Synoviolin 1) E3 ubiquitin ligase, component of the reticulum quality control system (ERAD) involved in ubiquitin dependent degradation of misfolded endoplasmic reticulum proteins 80 , 81 . RPS27A (Ribosomal Protein 27a) a fusion protein consisting of ubiquitin at the N terminus and the ribosomal protein 27a at the C-terminus], and RPS40 (Ribosomal Protein 40), implicated in maintenance of chromatin structure.
Compelling data support the role of SUMOylation in maturing oocytes, deletion of SUMO-component UBE2I disrupts meiotic maturation and causes defects in spindle organization 82 . Deletion of UBE2I impaired the communication with granulosa cells and caused a defective resumption of meiosis and meiotic progression 83 . As DAS gene, we also found SAE (SUMO1 Activating Enzyme Subunit 1) that mediates ATP-dependent activation of SUMO protein on a conserved cysteine residue on UBA2.
DAS genes from CCs GV-vs-M2 comprise several Small Ubiquitin-like modifier (SUMO) ligases: PIAS2 (Protein Inhibitor of activated STAT 2) and PIAS3 both stabilize the interaction with UBE2I and the substrate. PIAS2 play a crucial role as transcriptional co-regulator in the p53 pathway and the steroid hormone signalling pathway. PIAS3 directly binds to several transcription factors, blocking or enhancing their activity.
By comparing CCs GV-M2 we found several DAS E3 ubiquitin ligases: MDMD2, STUB1, HUWE1, HERC2, TRIM37, CBLB, CUL4B. Pathways regulated by TRIM (Tripartite Motif Containing) 37 are epigenetic transcriptional repression (mono-ubiquitination of histone H2A) 84 , centriole duplication 85 , peroxisome biogenesis 86 . While HERC2 (HECT and RLD Domain Containing Ubiquitin Protein Ligase 2) regulates small ubiquitin-dependent retention of repair proteins on damaged chromosome 87 . DDB2 (damage specific DNA binding protein 2) in complex with CUL4 may ubiquinate histones, facilitating their removal from nucleosome and promoting subsequent DNA repair 88 , 89 . HUWE1 (HECT E6AP type E3 ubiquitin protein ligase) targets the p53 tumour suppressor 90 , core histones 91 , 92 and DNA polymerase beta, playing a role in base-excision repair 93 . STUB1 (STIP1 Homology and U-Box containing Protein 1) mediates poly-ubiquitination of DNA polymerase beta (POLB), amplifying the HUWE1/ARF-BP1 dependent POLB-degradation by the proteasome 93 . In addition, STUB1 act as a co-chaperone for HSPA1A and HSPA1B chaperone proteins and promotes protein degradation 94 . CUL4B (Cullin 4B) is required for ubiquitination of cyclin E and consequently for normal G1 cell cycle progression 95 , 96 and for the proteolysis of several regulators of DNA replication. CUL4B regulates the mammalian target-of-rapamycin (mTOR) pathways implicated in the control of cell growth and metabolism 97 . Lastly, MDM2 (mouse double minute 2 homolog) a nuclear-localized E3 ubiquitin ligase, inhibits p53-mediated cell cycle arrest, promotes degradation of retinoblastoma RB1 protein 98 . Together these findings highlight that cumulus expansion is fundamental for oocyte maturation, as many DAS genes are involved in cell cycle control, DNA replication and repair.
Finally, comparison of CCs GV-M2 reveals multiple DAS genes encoding for spliceosome components. SRSF2, SRSF3, SRSF4, SRSF5, SRSF7 (Serine and Arginine Rich Splicing Factor 2, 3, 4, 5 and 7) are splicing factors that promote exon-inclusion during alternative splicing as well as mRNA nuclear export 99 , 100 , 101 . SNRPG (Small Nuclear Ribonucleoprotein Polypeptide G) is a component of precursors of the spliceosome like U1, U2, U4, U5 and U7 small nuclear ribonucleoprotein complexes. U5SNRNP70 (Small Nuclear Ribonucleoprotein U1 Subunit 70) is a component of the spliceosome U1 snRNP 102 , 103 . THOC2 (THO Complex Subunit 2) is a component of the TREX complex which is involved in mRNA processing and nuclear export.
PRPF38B (Pre-mRNA Processing Factor 38B), PRPF40A (Pre-mRNA Processing Factor 40 A) PRPF40B (Pre-mRNA Processing Factor 40B) are RNA binding protein involved in mRNA splicing and in several processes, including cytoskeleton organization, regulation of cell shape, regulation of cytokinesis. TRA2A (Transformer 2 Alpha Homolog) and TXNL4A (Tiredoxin like 4A) are RNA binding proteins involved in pre-mRNA splicing. RBM17 (RNA Binding Motif Protein 17) is a splice factor that binds to the single stranded 3′AG at the exon/intron border, also involved in the regulation of alternative splicing and the utilization of cryptic splice sites. NCBP2 (Nuclear Cap Binding Protein Subunit 2) is a component of cap-binding complex associated with pre-mRNA splicing, translational regulation, non-sense-mediated mRNA decay, RNA-mediated gene silencing by microRNAs and mRNA export. HNRNPA1 is a member of heterogeneous nuclear ribonucleoproteins (hnRNPS) associated with pre-mRNAs in the nucleus and pre-mRNAs packaging and processing. PQPB1 (Polyglutamine Binding Protein 1) is an intrinsically disordered protein that act as scaffold in different processes like pre-mRNA splicing, transcription regulation. Through its disordered region, PQPB1 regulates alternative splicing of specific pre-mRNAs molecules 104 , 105 , 106 , 107 . DHX38 (Dead- Box Helicase 38) is involved in pre-mRNA splicing as component of spliceosome. U2AF1 (U2 small Nuclear RNA A10uxiliary Factor 1) and U2AF1L4 (U2 small Nuclear RNA Auxiliary Factor 1 like 4) take part in protein-protein interactions and protein-RNA interactions required for an accurate 3′ splice site selection, while TCERG1 (Transcription Elongation Regulator 1) is a nuclear protein that regulates transcriptional elongation and pre-mRNA splicing.
In the last part of our study, we focused on miRNAs and their target genes. In fact, it is known that several miRNAs are intricately involved in the regulation of granulosa cell processes, crucial for ovarian function and folliculogenesis 108 , 109 .
Interestingly, by comparing the lists of DEMs shared by the two GC types, we found 12 DEMs showing a similar expression profile. A further GO analysis indicated significant common enriched terms associated with cell division, proliferation and metabolism, revealing a complex and powerful regulatory network within the two GC subpopulations.
Lastly, we built a co-expression miRNA-mRNA network to infer some potential regulatory relationships. A total of 12 common miRNAs were analyzed in the context of DE-mRNAs. In line with earlier studies, we found that miR-129-5p is the most connected in the miRNA-mRNA network 109 , while the two couples miR-30a-5p/miR-148a and miR-30a-3p/miR-196a-5p may act on common target genes. Interestingly, miR-129 is down-regulated in GCs of PCOS patients, whereas the overexpression of miR-129 increases P4 (Progesterone) and E2 (17-beta-estradiol) secretion, thus promoting GCs proliferation in PCOS 110 .
As previously reported, the miR-146a-5p expression, higher in MGCs compared to CCs, promotes apoptosis by targeting IRAK1 in MGCs 15 .
MiR-30a-3p and miR-30a-5p, belonging to the same miR-30 family, are both up-regulated in MGCs in contrast to miR-148a and miR-196-5p. The MiR-30 family is highly expressed in mammalian gonads, associated with the Homeobox protein and Zn transport 111 and with the regulation of ubiquitin-mediated pathways 112 . MiR-30a expression in ovarian cancer results in being significantly up-regulated 113 , 114 and further bioinformatics analysis indicates that miR-30a play a crucial role in ovarian cancer biology 115 . Alteration of miRNA expression profiles is associated with PCOS; miR-30a was detected in human follicular fluid and used as biomarkers (in tandem with miR-140 and let-7b) to discriminate between PCOS and normal ovarian reserve with a high specificity and sensitivity 116 .
Recent data show a significant dysregulation of miR-144 117 in ovarian tissues of animal model of PCOS and of miR-145 in granulosa cells 118 and miR-142 119 in follicular fluid, respectively, of PCOS patients.
Compelling evidence shows that miR-148a-3p modulates the Wnt/beta-catenin signaling pathway 120 and Serpin family E member 1 (SERPINE1) expression 121 .
In granulosa cells, SERPINE1 expression can influence follicular development by regulating matrix degradation and tissue remodeling processes necessary for follicle growth and ovulation. Aberrant expression of SERPINE1 can disrupt these processes, leading to reproductive issues such as PCOS 122 . Furthermore, miR-148a-3p is implicated in the regulation of steroidogenesis and cell proliferation within chicken granulosa cells, targeting genes vital for these processes 123 .
In addition, hsa-miR-196a-5p regulates granulosa cell apoptosis, affecting cell survival pathways crucial for follicular development by repressing Rho Associated Coiled-coil Containing Protein Kinase 1 (ROCK1) 124 . ROCK1, a key regulator of the actin cytoskeleton and cell polarity, ensures that apoptotic signals are properly localized within the cell, facilitating the efficient execution of the apoptotic program 125 .
MiR-145 targets Crkl and through the JNK/p38 MAPK pathway promotes cell proliferation, differentiation, and steroidogenesis 126 by regulating the cytoskeleton remodeling 127 .
The expression of miR-10b is higher in MGCs compared to CCs, while for miR-92a-1-5 it is the opposite. Recent data demonstrate that miR-10b represses BDNF expression in granulosa cells, by targeting the 3′ UTR 128 . The miR-10b expression is controlled by hormones and growth factors in the follicle, such as FSH, FGF9 and some ligands of the TGF-beta pathway. On the other hand, the miR-10 family inhibits many key genes of TGF-beta signaling axis, suggesting the existence of a negative feedback loop 129 . SMAD4 is a transcription factor of the CYP19A1 gene (encoding aromatase, a key enzyme in E2 synthesis), that in turn stimulates E2 release and inhibits cell apoptosis in porcine GCs. In contrast, miR-10b might act as a pro-apoptotic factor, directly interacting with 3′-UTR of porcine CYP19A1 mRNA, inhibiting its expression and function 130 . On the other hand, recent data demonstrate that miR-92a inhibits GCs apoptosis in pig ovaries, by targeting SMAD7 directly 131 . Very recent reports have highlighted the emerging role of miRNAs in regulating ovarian angiogenesis, follicular communication and in regulatory networks of cumulus-oocyte complex (COC) linked to fertilization 132 , 133 . The miRNA common network, herein identified, might likely work to fine tune various aspects of follicle maturation [i.e. cumulus cell survival/expansion, cumulus-oocyte communication, follicular microenvironment, angiogenesis]. Interestingly, miRNA profiling from human GC and follicular fluid has revealed the presence miRNAs, involved in the regulation of main pathways underlying folliculogenesis (like Wnt signaling 134 , mitogen-activated protein kinase 135 and phosphatidylinositol 3 kinase-protein kinase B 136 pathways respectively). Among these miRNAs, miRNA-10b-5p that we found upregulated in MGCs 137 . Furthermore, analysis of the follicular fluid from bovine ovaries unveiled a role of few miRNAs in global DNA demethylation 138 . By targeting DNMT, miRNA-148a that we found upregulated in MGCs, may impact on DNA methylation, thus improving embryonic development 138 . MiRNA profiling of exosomes derived either from follicular fluid (FF) or from human GCs seem to show a remarkable overlap 137 . Our study goes in the same direction, since we have identified a shared miRNA signature within GCs. Emerging in-depth analysis of miRNA profiles from exosomes (derived from FF and GCs) will allow soon to better define the miRNA signature and its positive effect on follicle development and activation.
In conclusion, these results have overcome the limitations of individual studies, enabling the identification of new candidates, genes, miRNAs, and signaling routes within the mature follicle. Such new molecular signatures can be used to study the mechanisms underlying GCs function, and as potential biomarkers of oocyte quality.
Data source
RNA-seq and small RNA-seq datasets related to granulosa cells and oocyte were retrieved by consulting the Sequence Read Archive (SRA) according to Search and Incorporation criteria detailed below.
Search criteria
A systematic search strategy was employed to identify relevant datasets in the SRA archive. The search was performed using keywords such as “granulosa cells”, “cumulus cells and oocyte” and “mural and granulosa cells” ensuring the specificity of the results. Then a filter for homo sapiens species was applied. The SRA archive’s web interface or command-line tools (e.g., SRA Toolkit) were utilized to execute the search queries. The search queries were constructed to include the relevant keywords and any additional filters or parameters deemed necessary.
Incorporation criteria
The identified datasets were evaluated based on specific selection criteria to ensure their suitability for the research project. Criteria considered may include data quality, experimental design, sample size (at least three replicates), tissue source, and relevant metadata. Only standard RNA-seq data was considered, single cell dataset was excluded. An important criterion was the platform of sequencing, indeed only sequencing data from the Illumina platform were chosen to ensure that the same pipeline could be set up and applied to the selected Bioprojects to harmonize all datasets before executing downstream integrated analyses.
Data selection
Three bioprojects of interest have been found in the Sequence Read Archive (SRA) for mRNA sequencing experiments on human granulosa cells, and two bioprojects for small RNA sequencing experiments have been found to evaluate the miRNAs profile of these cells. Data selection was focused on experiments that included also the cumulus cells (CCs).
Briefly, from the article by Yerushalmi et al . 12 were analyzed CCs in two stages to get insight into different maturation phases of the COC, it was linked to PRJNA216966, which has 2 samples of CCs from germinal vesicles unstimulated COC, and 3 samples from M2 cumulus of expanded/stimulated COC.
Li et al . 13 focused on Polycystic Ovary Syndrome. To discover what characterizes cumulus cells from oocytes and find new insight into intercellular communication, the analysis was only performed considering the control groups. From PRJNA649934, 6 control oocyte samples and 4 control cumulus cells samples.
In the article by Velthut-Meikas et al . 14 , two bioprojects were linked PRJNA200696 and PRJNA200699 respectively RNA-seq and small RNA-seq experiments on Cumulus and Mural cells in the preovulatory stage. All samples were treated with the standard stimulation protocol: GnRH antagonist, rFSH, hCG. The former has 6 samples total, 3 for CCs and 3 for mural cells (MCs), the latter has 12 samples, 6 for CCs and 6 for MCs.
Completing the small RNA-seq data on granulosa cells is PRJNA417973 15 . 24 runs total, 12 for CCs (3 samples, 4 runs per sample) and 12 for MCs (3 samples) have been analyzed.
In our analysis, we observed a substantial disproportion in the number of PCOS datasets compared to control and standard treatment datasets. We did not prioritize the analysis of PCOS datasets (like PRJNA762274, PRJEB46048, PRJNA576435, PRJNA576231) since our primary objective was to investigate and characterize the transcriptome of Cumulus Cells under control conditions. To mitigate the inherent bias caused by the unequal distribution of datasets, we employed stringent criteria for dataset selection. We prioritized datasets with well-defined metadata, rigorous experimental design, and a balanced representation of PCOS cases, control subjects, and standard treatment groups. Datasets lacking essential information or exhibiting a skewed distribution were regrettably excluded from our analysis to ensure the robustness and validity of our findings.
Data download
The selected datasets were downloaded from the SRA archive using SRA toolkit v3.0.0. The chosen dataset’s accession numbers or unique identifiers were recorded for future reference. The whole metadata are reported under supplementary table 1 .
mRNA-Seq data processing
Quality control and trimming.
A first quality control was performed on the FASTQ files using FASTQC (version 0.11.9), low-quality reads and adapters were removed using Trimmomatic v.0.39 (installed in g100). For adapter trimming were used the TrueSeq adapters for paired end:
>PrefixPE/1
TACACTCTTTCCCTACACGACGCTCTTCCGATCT
>PrefixPE/2
GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT
AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTA
AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC
For single end:
>TruSeq 3_IndexedAdapter
>TruSeq 3_UniversalAdapter
Parameters used for trimmomatic for paired-end and single-end reads were LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36.
Alignment and annotation
For the alignment, The HISAT2 aligner software (version 2.2.1) 139 was employed for aligning the RNA-Seq reads to the reference genome assembly for Homo Sapiens (hg38). Prior to alignment, the reference genome assembly was indexed using HISAT2’s indexing utility. The alignment of RNA-Seq reads to the reference genome was performed using HISAT2 principal command. Were used both paired and unpaired data after trimming.
The resulting alignment file in SAM format was converted to the binary BAM format for efficient storage and subsequent analysis and to facilitate downstream analysis, the BAM file was sorted by genomic coordinates. The conversion was performed using the SAMtools (version 1.13) 140 .
If the experiment had multiple runs for the same sample, the BAM files were merged using SAMtools. As a result, there was a BAM file for each sample.
The quantification of gene expression levels based on the aligned reads and a genome annotation file (GTF or GFF) was performed using Stringtie (version 2.1.6) 141 . This step involved assigning reads to genes and counting the number of reads associated with each gene. The genome annotation file was obtained from hg38 19 release and provided in GTF format. The PrepDe python script was used to generate the resulting gene count matrix for differential expression analysis, as described in the StringTie handbook 142 .
Data quality control before dataset integration
The datasets coming from different bioprojects have been processed using the shared pipeline as described above. This consistent processing helps mitigating some of the variability introduced by differences in library preparation and sequencing depth. However, we recognize that differences in library preparation and sequencing depth can influence gene detection efficiency and expression levels, potentially affecting downstream pathway analysis. To address this, we have conducted several quality control measures to assess and account for these differences.
To provide a clearer understanding of the dataset quality and sequencing depth, we have conducted a fastqc analysis, which is reported under the Supplementary Dataset 143 . The MultiQC report provides a comprehensive overview of the raw fastq data quality, including metrics such as read quality scores, adapter content, and duplication levels before and after trimming. Furthermore, we have conducted a PCA on the count matrices for each bioproject (S7). This analysis helps visualizing the similarity and differences in gene expression profiles. The PCA analysis has clearly highlighted distinct sample clustering at dataset level segregating them by cell type and not by potentially confounding batch effects. The subsequent gene-count normalization process operated by DESeq 2 package ensured proper comparisons and a robust final list of DEGs.
mRNA post-processing data analysis
Most post-processing analysis were conducted using R programming language with R studio (version 2022.02.3 Build 492). The raw count data obtained from Stringtie was used as input for differential expression analysis. The DESeq 2 R package (version 1.36.0) 144 was used to normalize the raw counts, estimate size factors, and perform statistical tests to identify differentially expressed genes. This analysis aimed to identify genes with significantly different expression levels between experimental conditions or cell types.
Statistical analysis was conducted using DESeq 2. The analysis considered relevant factors such as treatment condition, replicates, and experimental design. Significantly differentially expressed genes were determined based on at least 0.05 adjusted p-values and two-log2 fold change thresholds in up- or down-regulation.
Gene Ontology (GO) enrichment analysis was performed using the clusterProfiler R package (version 4.7.1.003) 145 to gain insights into the functional annotation and enrichment of DEGs within bioprojects. The gene list used for GO analysis consisted of genes that showed significant differential expression both up- or downregulated between experimental conditions or cell types, as determined by the differential expression analysis using DESeq 2.
We acknowledge that DEGs from two of the three datasets might include markers specific to mural GCs and oocytes, which could influence the functional analysis results. To address this, we conducted a detailed analysis to show the up-regulated and down-regulated genes separately in each cell type across multiple datasets (Fig. S4 ).
Before conducting GO-analysis, if necessary, the gene identifiers in the input gene list were converted to a common identifier system, such as Entrez Gene IDs or official gene symbols, using available annotation database org.Hs.eg.db 146 .
The gene list was annotated with GO terms using the enrichGO function from clusterProfiler. The annotation was based on the specific organism’s GO database (org.Hs.eg.db). The enrichGO function was used to identify significantly enriched GO terms within the gene list. This function performed a hypergeometric test to determine if a particular GO term was overrepresented in the input gene list compared to the genome background. The p-values were adjusted for multiple testing using the appropriate method Benjamini-Hochberg correction method.
The results of the GO enrichment analysis were visualized using various plotting functions from clusterProfiler. To remove redundancy of enriched GO terms the simplify function from the clusterProfiler package. This step provided a more concise and interpretable representation of the enriched GO terms.
To perform KEGG pathway enrichment analysis the clusterProfiler package was used following the same steps of the GO enrichment analysis except for the database was downloaded locally due to known problems with clusterProfiler connecting to KEGG DB, loading the “KEGG.db” downloaded from the create_kegg_db(“hsa”) command, using the “createKEGGdb” R package.
In parallel, to reveal the hidden patterns of common biological processes, the R package clusterProfiler was also used in compareCluster modality, where the complete lists of DEGs from each Bioproject are used simultaneously as input.
Protein Protein Interaction network was built using the webAPP STRING v12.0 using the coreset of 236 shared DEGs as input.
Differential alternative splicing analysis
To investigate differential alternative splicing events between experimental conditions or treatments, MAJIQ (version 2.4) was employed 147 , 148 . The MAJIQ software was downloaded from the official website 149 and installed via Conda. The required dependencies and reference annotations were obtained according to the MAJIQ documentation. A MAJIQ configuration file was created to specify the experimental design and sample information. This file included details such as the input BAM files for each condition or treatment, sample groupings, and experimental factors.
MAJIQ “build” command was used to detect and quantify alternative splicing events from the aligned RNA-Seq data in BAM format. Furthermore, a gene annotation file in gff3 format was required, the file used for the analysis is available on genecode. The output is a.majiq file for each sample, which is used as input for the “deltapsi” command to identify differentially spliced events between conditions or cell type.
The output of the differential splicing analysis provided the differential splicing events, including their psi values (percent spliced-in) and statistical significance.
MAJIQ calculates differential alternative splicing over local splicing variations (LSVs), not on the entire gene. These results make it so that even small LSVs variations (20%) can strongly affect the differential splicing, thus this filtering criterion, in conjunction with q < 0.05, was used to produce filtered lists of LSVs.
Various visualization tools and packages, such as Voila: a visualization package that combines the output of MAJIQ Builder (“build”) and MAJIQ Quantifier(“deltapsi”) using interactive D3 components and HTML5. Then the Modulizer tool was used to categorize different splicing event types. To get the distribution of each alternative splicing category in each Bioproject, a custom script was built.
To gain insights into the biological functions and pathways associated with the differentially alternative splicing (DAS), GO Enrichment Analysis was performed on DAS transcription regulators. The DAS genes can be linked to their corresponding gene annotations or functional databases to determine enriched terms or pathways. The enrichGO function was used to identify significantly enriched GO terms within the gene list. Additional analysis on transcription regulator DAS and their targets was performed using the TRRUST database. Targets were filtered to only the differentially expressed genes, then a GO enrichment analysis was performed on the filtered target list 150 .
The violin plot implemented in the Fig. S4 serves as an exploratory visualization tool. Its primary input consists of the lists of DEGs associated with each TF within individual bioprojects. The plot is further annotated to identify instances where genes exhibit both being TF and DAS. This visualization is created using the GGplot2 R package.
Small RNA-seq analysis
For PRJNA200699 was used miRDeep2 (version 0.1.2) 151 to trim, detect and quantify miRNAs expression.
For PRJNA417973 a specific adapter trimming was required due to the library construction 152 , Cutadapt v4.2 was employed with the following commands: cutadapt–discard-untrimmed–minimum-length = 18–maximum-length = 35 -a NNNNTGGAATTCTCGGGTGCCAAGGNNNN -o clipped_SAMN08013076.fastq -j 0 -O 11 SAMN08013076.fastq.
Gene ontology enrichment analysis was performed as previously described the gene list for the enrichment is the list of differential expressed miRNAs target genes (experimentally validated only). Then a venn diagram has been plotted to represent the shared DEMs between the two bioprojects using ggVenn from ggplot2 and VennDiagram R package 153 .
An interaction network was built selecting differentially expressed miRNAs shared in both projects and their target DEGs from PRJNA200696 using igraph R package, and Fruchterman-Reingold algorithm for graph visualization. miRNA-targets were downloaded using multiMiR R package 154 , applying the validated target filter.
List of abbreviations and acronyms used in the paper
GCs (Granulosa Cells); CCs (Cumulus Cells); miRNAs (microRNAs); mRNAs (messenger RNAs); DEM (Differentially expressed miRNAs); DEGs (Differentially expressed Genes); DAS (Differential alternative splicing); cumulus-oocyte complex (COC); human chorionic gonadotrophin (hCG); metaphase 2 (M2); Polycystic Ovary syndrome (PCOS); Differentially Expressed (DE); Sequence Read Archive (SRA); Gene Ontology (GO); Kyoto Encyclopedia of Genes and Genomes (KEGG); transcription regulator (TR); ALE/AFE (Alternative Last/First Exon); MES (Multiple Exon skipping); MXE (Mutually Exclusive Exons); alt3′ (alternative 3′ exon); alt5′ (alternative 5′ exon).
Data availability
Normalized gene expression count tables, DEGs/DEMs/DAS lists and any other relevant sample metadata were deposited on the repository Figshare 17 , 73 , 143 , 155 .
Code availability
The workflow and a detailed list of command lines was deposited on the repository https://github.com/Xhulio-99/RNAseq_analysis_code_for_human_granulosa_cells_paper .
Li, R. & Albertini, D. F. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell Biol. 14 , 141–152 (2013).
Article CAS PubMed Google Scholar
Turathum, B., Gao, E. M. & Chian, R. C. The Function of Cumulus Cells in Oocyte Growth and Maturation and in Subsequent Ovulation and Fertilization. Cells 10 (2021).
Da Broi, M. G. et al . Influence of follicular fluid and cumulus cells on oocyte quality: clinical implications. J. Assist. Reprod. Genet. 35 , 735–751 (2018).
Article PubMed PubMed Central Google Scholar
Dumesic, D. A., Meldrum, D. R., Katz-Jaffe, M. G., Krisher, R. L. & Schoolcraft, W. B. Oocyte environment: follicular fluid and cumulus cells are critical for oocyte health. Fertil. Steril. 103 , 303–316 (2015).
Article PubMed Google Scholar
Grøndahl, M. L. et al . Specific genes are selectively expressed between cumulus and granulosa cells from individual human pre-ovulatory follicles. Mol. Hum. Reprod. 18 , 572–584 (2012).
Khan, D. R. et al . Meta-analysis of gene expression profiles in granulosa cells during folliculogenesis. Reproduction 151 , R103–R110 (2016).
Wigglesworth, K., Lee, K. B., Emori, C., Sugiura, K. & Eppig, J. J. Transcriptomic diversification of developing cumulus and mural granulosa cells in mouse ovarian follicles. Biol. Reprod . 92 (2015).
Uyar, A., Torrealday, S. & Seli, E. Cumulus and granulosa cell markers of oocyte and embryo quality. Fertil. Steril. 99 , 979–997 (2013).
Ouandaogo, Z. G. et al . Differences in transcriptomic profiles of human cumulus cells isolated from oocytes at GV, MI and MII stages after in vivo and in vitro oocyte maturation. Hum. Reprod. 27 , 2438–2447 (2012).
Péquignot, Y. & Clarke, P. G. H. Reduction in the death and cytolamination of developing neurons by tetrodotoxin in the axonal target region. Brain Res. Dev. Brain Res. 60 , 94–98 (1991).
De Los Santos, M. J. et al . Hormonal and molecular characterization of follicular fluid, cumulus cells and oocytes from pre-ovulatory follicles in stimulated and unstimulated cycles. Hum. Reprod. 27 , 1596–1605 (2012).
Article Google Scholar
Yerushalmi, G. M. et al . Characterization of the human cumulus cell transcriptome during final follicular maturation and ovulation. Mol. Hum. Reprod. 20 , 719–735 (2014).
Li, J. et al . Molecular Features of Polycystic Ovary Syndrome Revealed by Transcriptome Analysis of Oocytes and Cumulus Cells. Front. cell Dev. Biol . 9 (2021).
Velthut-Meikas, A. et al . Research resource: small RNA-seq of human granulosa cells reveals miRNAs in FSHR and aromatase genes. Mol. Endocrinol. 27 , 1128–1141 (2013).
Article CAS PubMed PubMed Central Google Scholar
Andrei, D. et al . Differential miRNA Expression Profiles in Cumulus and Mural Granulosa Cells from Human Pre-ovulatory Follicles. MicroRNA (Shariqah, United Arab Emirates) 8 , 61–67 (2019).
CAS PubMed Google Scholar
Home - SRA - NCBI. https://www.ncbi.nlm.nih.gov/sra .
Dhori, X. Gioiosa, S. & Gonfloni, S. mRNA Analysis (Gene counts, DEGs, GOs, KEGGs) . Figshare https://doi.org/10.6084/m9.figshare.23816205.v2 (2024).
Szklarczyk, D. et al . The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51 , D638–D646 (2023).
Duan, X. & Sun, S. C. Actin cytoskeleton dynamics in mammalian oocyte meiosis. Biol. Reprod. 100 , 15–24 (2019).
Chakrabarti, R., Lee, M. & Higgs, H. N. Multiple roles for actin in secretory and endocytic pathways. Curr. Biol. 31 , R603–R618 (2021).
Gunning, P., O’Neill, G. & Hardeman, E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 88 , 1–35 (2008).
Rasmussen, C. G., Wright, A. J. & Müller, S. The role of the cytoskeleton and associated proteins in determination of the plant cell division plane. Plant J. 75 , 258–269 (2013).
Baumgarten, S. C. et al . IGF1R signaling is necessary for FSH-induced activation of AKT and differentiation of human Cumulus granulosa cells. J. Clin. Endocrinol. Metab. 99 , 2995–3004 (2014).
Stocco, C. et al . Genome-wide interactions between FSH and insulin-like growth factors in the regulation of human granulosa cell differentiation. Hum. Reprod. 32 , 905–914 (2017).
Huet, C., Monget, P., Pisselet, C. & Monniaux, D. Changes in extracellular matrix components and steroidogenic enzymes during growth and atresia of antral ovarian follicles in the sheep. Biol. Reprod. 56 , 1025–1034 (1997).
Huet, C. et al . Chronology of events accompanying follicular atresia in hypophysectomized ewes. Changes in levels of steroidogenic enzymes, connexin 43, insulin-like growth factor II/mannose 6 phosphate receptor, extracellular matrix components, and matrix metalloproteinases. Biol. Reprod. 58 , 175–185 (1998).
Huet, C., Pisselet, C., Mandon-Pépin, B., Monget, P. & Monniaux, D. Extracellular matrix regulates ovine granulosa cell survival, proliferation and steroidogenesis: relationships between cell shape and function. J. Endocrinol. 169 , 347–360 (2001).
Berkholtz, C. B., Lai, B. E., Woodruff, T. K. & Shea, L. D. Distribution of extracellular matrix proteins type I collagen, type IV collagen, fibronectin, and laminin in mouse folliculogenesis. Histochem. Cell Biol. 126 , 583–592 (2006).
Diaz, F. J., Wigglesworth, K. & Eppig, J. J. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J. Cell Sci. 120 , 1330–1340 (2007).
Eppig, J. J. Intercommunication between mammalian oocytes and companion somatic cells. Bioessays 13 , 569–574 (1991).
Richards, J. A. S., Russell, D. L., Ochsner, S. & Espey, L. L. Ovulation: new dimensions and new regulators of the inflammatory-like response. Annu. Rev. Physiol. 64 , 69–92 (2002).
Eppig, J. J., Pendola, F. L., Wigglesworth, K. & Pendola, J. K. Mouse oocytes regulate metabolic cooperativity between granulosa cells and oocytes: amino acid transport. Biol. Reprod. 73 , 351–357 (2005).
Hu, Y. C. et al . Subfertility and defective folliculogenesis in female mice lacking androgen receptor. Proc. Natl. Acad. Sci. USA 101 , 11209–11214 (2004).
Article ADS CAS PubMed PubMed Central Google Scholar
Van Soom, A., Tanghe, S., De Pauw, I., Maes, D. & De Kruif, A. Function of the cumulus oophorus before and during mammalian fertilization. Reprod. Domest. Anim. 37 , 144–151 (2002).
Guzeloglu-Kayisli, O. et al . Embryonic poly(A)-binding protein (EPAB) is required for oocyte maturation and female fertility in mice. Biochem. J. 446 , 47–58 (2012).
Schellander, K., Hoelker, M. & Tesfaye, D. Selective degradation of transcripts in mammalian oocytes and embryos. Theriogenology 68 Suppl 1 (2007).
Massey, P. Intravenous nutrient therapy eliminated androgen deprivation therapy-induced hot flashes in two men with prostate cancer. J. Soc. Integr. Oncol. 4 , 153–156 (2006).
Walser, C. B. & Lipshitz, H. D. Transcript clearance during the maternal-to-zygotic transition. Curr. Opin. Genet. Dev. 21 , 431–443 (2011).
Yu, C. et al . Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res. 26 , 275–287 (2016).
Sirard, M. A. Resumption of meiosis: mechanism involved in meiotic progression and its relation with developmental competence. Theriogenology 55 , 1241–1254 (2001).
Chen, J. et al . Somatic cells regulate maternal mRNA translation and developmental competence of mouse oocytes. Nat. Cell Biol. 15 , 1415–1423 (2013).
Pan, Z. N. et al . RAB7 GTPase regulates actin dynamics for DRP1-mediated mitochondria function and spindle migration in mouse oocyte meiosis. FASEB J. 34 , 9615–9627 (2020).
Sun, M. H. et al . Citrinin exposure disrupts organelle distribution and functions in mouse oocytes. Environ. Res . 185 (2020).
Xu, Y. et al . Nonylphenol exposure affects mouse oocyte quality by inducing spindle defects and mitochondria dysfunction. Environ. Pollut . 266 (2020).
Van Blerkom, J. Mitochondria in human oogenesis and preimplantation embryogenesis: engines of metabolism, ionic regulation and developmental competence. Reproduction 128 , 269–280 (2004).
Dumollard, R., Duchen, M. & Sardet, C. Calcium signals and mitochondria at fertilisation. Semin. Cell Dev. Biol. 17 , 314–323 (2006).
Liu, M., Sims, D. A., Calarco, P. & Talbot, P. Biochemical heterogeneity, migration, and pre-fertilization release of mouse oocyte cortical granules. Reprod. Biol. Endocrinol . 1 (2003).
Liu, M. The biology and dynamics of mammalian cortical granules. Reprod. Biol. Endocrinol . 9 (2011).
Verlhac, M. H., Lefebvre, C., Guillaud, P., Rassinier, P. & Maro, B. Asymmetric division in mouse oocytes: with or without Mos. Curr. Biol. 10 , 1303–1306 (2000).
Machaca, K. Ca2+ signaling differentiation during oocyte maturation. J. Cell. Physiol. 213 , 331–340 (2007).
FitzHarris, G., Marangos, P. & Carroll, J. Changes in endoplasmic reticulum structure during mouse oocyte maturation are controlled by the cytoskeleton and cytoplasmic dynein. Dev. Biol. 305 , 133–144 (2007).
Kan, R. et al . Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Dev. Biol. 350 , 311–322 (2011).
Sun, M. H. et al . Ral GTPase is essential for actin dynamics and Golgi apparatus distribution in mouse oocyte maturation. Cell Div . 16 (2021).
He, M., Zhang, T., Yang, Y. & Wang, C. Mechanisms of Oocyte Maturation and Related Epigenetic Regulation. Front. cell Dev. Biol . 9 (2021).
Arora, A. et al . The Role of Alternative Polyadenylation in the Regulation of Subcellular RNA Localization. Front. Genet . 12 (2022).
Piccioni, F., Zappavigna, V. & Verrotti, A. C. Translational regulation during oogenesis and early development: the cap-poly(A) tail relationship. C. R. Biol. 328 , 863–881 (2005).
Chen, J. et al . Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 25 , 755–766 (2011).
Su, Y. Q. et al . Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev. Biol. 302 , 104–117 (2007).
Sha, Q. Q., Zhang, J. & Fan, H. Y. A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals†. Biol. Reprod. 101 , 579–590 (2019).
Yang, Y. et al . Maternal mRNAs with distinct 3′ UTRs define the temporal pattern of Ccnb1 synthesis during mouse oocyte meiotic maturation. Genes Dev. 31 , 1302–1307 (2017).
Bennett, J., Baumgarten, S. C. & Stocco, C. GATA4 and GATA6 silencing in ovarian granulosa cells affects levels of mRNAs involved in steroidogenesis, extracellular structure organization, IGF-I activity, and apoptosis. Endocrinology 154 , 4845–4858 (2013).
Rotgers, E. et al . Constitutive expression of Steroidogenic factor-1 (NR5A1) disrupts ovarian functions, fertility, and metabolic homeostasis in female mice. FASEB J . 35 (2021).
Suyama, A. et al . Roles of NR5A1 and NR5A2 in the regulation of steroidogenesis by Clock gene and bone morphogenetic proteins by human granulosa cells. Endocr. J. 68 , 1283–1291 (2021).
Jeyasuria, P. et al . Cell-specific knockout of steroidogenic factor 1 reveals its essential roles in gonadal function. Mol. Endocrinol. 18 , 1610–1619 (2004).
Halbleib, J. M. & Nelson, W. J. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20 , 3199–3214 (2006).
Maître, J. L. & Heisenberg, C. P. Three functions of cadherins in cell adhesion. Curr. Biol . 23 (2013).
St. Croix, B. et al . E-Cadherin-dependent growth suppression is mediated by the cyclin-dependent kinase inhibitor p27(KIP1). J. Cell Biol. 142 , 557–571 (1998).
Klezovitch, O. & Vasioukhin, V. Cadherin signaling: Keeping cells in touch. F1000Research 4 (2015).
Kim, N. G., Koh, E., Chen, X. & Gumbiner, B. M. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. USA 108 , 11930–11935 (2011).
Silvis, M. R. et al . α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci. Signal . 4 (2011).
Stepniak, E., Radice, G. L. & Vasioukhin, V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb. Perspect. Biol . 1 (2009).
Piprek, R. P. Molecular mechanisms underlying female sex determination–antagonism between female and male pathway. Folia Biol. (Praha). 57 , 105–113 (2009).
Article CAS Google Scholar
Dhori, X., Gioiosa, S. & Gonfloni, S. Alternative Splicing dataset. Figshare https://doi.org/10.6084/m9.figshare.23822445.v2 (2024).
Molecular Cloning of a Novel Member of the Eukaryotic Polypeptide Chain-Releasing Factors (eRF). Journal of Biological Chemistry 273 (35), 22254–22259 https://doi.org/10.1074/jbc.273.35.22254 (1998).
Bassermann, F., Eichner, R. & Pagano, M. The ubiquitin proteasome system - implications for cell cycle control and the targeted treatment of cancer. Biochim. Biophys. Acta 1843 , 150–162 (2014).
Jones, K. T. Anaphase-promoting complex control in female mouse meiosis. Results Probl. Cell Differ. 53 , 343–363 (2011).
Huo, L. J. et al . Regulation of ubiquitin-proteasome pathway on pig oocyte meiotic maturation and fertilization. Biol. Reprod. 71 , 853–862 (2004).
Sato, T. et al . STT3B-dependent posttranslational N-glycosylation as a surveillance system for secretory protein. Mol. Cell 47 , 99–110 (2012).
Jo, Y., Hartman, I. Z. & DeBose-Boyd, R. A. Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol. Biol. Cell 24 , 169–183 (2013).
Kaneko, M., Ishiguro, M., Niinuma, Y., Uesugi, M. & Nomura, Y. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 532 , 147–152 (2002).
Nadav, E. et al . A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem. Biophys. Res. Commun. 303 , 91–97 (2003).
Yuan, Y. F. et al . SUMO-1 plays crucial roles for spindle organization, chromosome congression, and chromosome segregation during mouse oocyte meiotic maturation. Mol. Reprod. Dev. 81 , 712–724 (2014).
Rodriguez, A. et al . Loss of the E2 SUMO-conjugating enzyme Ube2i in oocytes during ovarian folliculogenesis causes infertility in mice. Development 146 (2019).
Bhatnagar, S. et al . TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature 516 , 116–120 (2014).
Balestra, F. R., Strnad, P., Flückiger, I. & Gönczy, P. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev. Cell 25 , 555–571 (2013).
Wang, W., Xia, Z. J., Farré, J. C. & Subramani, S. TRIM37, a novel E3 ligase for PEX5-mediated peroxisomal matrix protein import. J. Cell Biol. 216 , 2843–2858 (2017).
Danielsen, J. R. et al . DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J. Cell Biol. 197 , 179–187 (2012).
Wang, H. et al . Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22 , 383–394 (2006).
Kapetanaki, M. G. et al . The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA 103 , 2588–2593 (2006).
Article ADS CAS PubMed Google Scholar
Yoon, S. Y. et al . Over-expression of human UREB1 in colorectal cancer: HECT domain of human UREB1 inhibits the activity of tumor suppressor p53 protein. Biochem. Biophys. Res. Commun. 326 , 7–17 (2005).
Liu, Z., Oughtred, R. & Wing, S. S. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol. Cell. Biol. 25 , 2819–2831 (2005).
Chen, D. et al . ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121 , 1071–1083 (2005).
Parsons, J. L. et al . Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J. 28 , 3207–3215 (2009).
Rajarajan, P., Gil, S. E., Brennand, K. J. & Akbarian, S. Spatial genome organization and cognition. Nat. Rev. Neurosci. 17 , 681–691 (2016).
Higa, L. A. et al . Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle 5 , 71–77 (2006).
Zou, Y. et al . Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J. Biol. Chem. 284 , 33320–33332 (2009).
Ghosh, P., Wu, M., Zhang, H. & Sun, H. mTORC1 signaling requires proteasomal function and the involvement of CUL4-DDB1 ubiquitin E3 ligase. Cell Cycle 7 , 373–381 (2008).
Hu, L., Zhang, H., Bergholz, J., Sun, S. & Xiao, Z. X. J. MDM2/MDMX: Master negative regulators for p53 and RB. Mol. Cell. Oncol . 3 (2016).
Animal welfare problems arising as a result of FMD: short-term solutions - PubMed. https://pubmed.ncbi.nlm.nih.gov/11338712/ .
Hautbergue, G. M., Hung, M. L., Golovanov, A. P., Lian, L. Y. & Wilson, S. A. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. USA 105 , 5154–5159 (2008).
Roundtree, I. A. et al . YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 6 (2017).
Pomeranz Krummel, D. A., Oubridge, C., Leung, A. K. W., Li, J. & Nagai, K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458 , 475–480 (2009).
Kondo, Y., Oubridge, C., van Roon, A. M. M. & Nagai, K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife 4 (2015).
Waragai, M. et al . PQBP-1, a novel polyglutamine tract-binding protein, inhibits transcription activation by Brn-2 and affects cell survival. Hum. Mol. Genet. 8 , 977–987 (1999).
Okazawa, H. et al . Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron 34 , 701–713 (2002).
Wang, Q., Moore, M. J., Adelmant, G., Marto, J. A. & Silver, P. A. PQBP1, a factor linked to intellectual disability, affects alternative splicing associated with neurite outgrowth. Genes Dev. 27 , 615–626 (2013).
Tapia, V. E. et al . Y65C missense mutation in the WW domain of the Golabi-Ito-Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA. splicing. J. Biol. Chem. 285 , 19391–19401 (2010).
Zhang, B. et al . MicroRNA Mediating Networks in Granulosa Cells Associated with Ovarian Follicular Development. Biomed Res. Int . 2017 (2017).
Gong, Z., Yang, J., Bai, S. & Wei, S. MicroRNAs regulate granulosa cells apoptosis and follicular development - A review. Asian-Australasian . J. Anim. Sci. 33 , 1714–1724 (2020).
CAS Google Scholar
Zhu, H. L., Chen, Y. Q. & Zhang, Z. F. Downregulation of lncRNA ZFAS1 and upregulation of microRNA-129 repress endocrine disturbance, increase proliferation and inhibit apoptosis of ovarian granulosa cells in polycystic ovarian syndrome by downregulating HMGB1. Genomics 112 , 3597–3608 (2020).
Madison-Villar, M. J. & Michalak, P. Misexpression of testicular microRNA in sterile Xenopus hybrids points to tetrapod-specific microRNAs associated with male fertility. J. Mol. Evol. 73 , 316–324 (2011).
Hadden, J. W. Steroid hormones and cancer. Clin. Bull. 10 , 121–126 (1980).
Wang, L., Zhao, S. & Yu, M. Mechanism of Low Expression of miR-30a-5p on Epithelial-Mesenchymal Transition and Metastasis in Ovarian Cancer. DNA Cell Biol. 38 , 341–351 (2019).
Wang, Y. et al . FOXD1 is targeted by miR-30a-5p and miR-200a-5p and suppresses the proliferation of human ovarian carcinoma cells by promoting p21 expression in a p53-independent manner. Int. J. Oncol. 52 , 2130–2142 (2018).
Lu, W. et al . Bioinformatics analysis of prognostic value and prospective pathway signal of miR-30a in ovarian cancer. J. Ovarian Res . 13 (2020).
Scalici, E. et al . Circulating microRNAs in follicular fluid, powerful tools to explore in vitro fertilization process. Sci. Rep . 6 (2016).
Xu, X., Xu, X., Wang, X. & Shen, L. Baicalin suppress the development of polycystic ovary syndrome via regulating the miR-874-3p/FOXO3 and miR-144/FOXO1 axis. Pharm. Biol. 61 , 878–885 (2023).
Naji, M. et al . Expression of miR-15a, miR-145, and miR-182 in granulosa-lutein cells, follicular fluid, and serum of women with polycystic ovary syndrome (PCOS). Arch. Gynecol. Obstet. 297 , 221–231 (2018).
Li, Y. et al . Dysregulated miR-142, -33b and -423 in granulosa cells target TGFBR1 and SMAD7: a possible role in polycystic ovary syndrome. Mol. Hum. Reprod. 25 , 638–646 (2019).
Ashmawy, A. M. et al . Crosstalk between liver-related microRNAs and Wnt/β-catenin pathway in hepatocellular carcinoma patients. Arab J. Gastroenterol. 18 , 144–150 (2017).
Hu, B. et al . MicroRNA-148a-3p Directly Targets SERPINE1 to Suppress EMT-Mediated Colon Adenocarcinoma Progression. Cancer Manag. Res. 13 , 6349–6362 (2021).
Bai, L. et al . Aberrant elevation of GDF8 impairs granulosa cell glucose metabolism via upregulating SERPINE1 expression in patients with PCOS. Mol. Ther. Nucleic Acids 23 , 294–309 (2020).
Xu, Z. et al . miRNA profiling of chicken follicles during follicular development. Sci. Rep . 14 (2024).
Chen, P. et al . MiR196a-5p in extracellular vesicles released from human nasopharyngeal carcinoma enhance the phagocytosis and secretion of microglia by targeting ROCK1. Exp. Cell Res . 411 (2022).
Sebbagh, M. et al . Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat. Cell Biol. 3 , 346–352 (2001).
Wang, S. et al . MiR-145 regulates steroidogenesis in mouse primary granulosa cells through targeting Crkl. Life Sci . 282 (2021).
Ma, L. et al . MiR-145 regulates steroidogenesis in mouse primary granulosa cells by targeting Arpc5 and subsequent cytoskeleton remodeling. J. Reprod. Dev. 69 , 154–162 (2023).
Peng, J. Y. et al . MicroRNA-10b suppresses goat granulosa cell proliferation by targeting brain-derived neurotropic factor. Domest. Anim. Endocrinol. 54 , 60–67 (2016).
Jiajie, T., Yanzhou, Y., Hoi-Hung, A. C., Chen, Z. J. & Wai-Yee, C. Conserved miR-10 family represses proliferation and induces apoptosis in ovarian granulosa cells. Sci. Rep . 7 (2017).
Li, Q., Du, X., Pan, Z., Zhang, L. & Li, Q. The transcription factor SMAD4 and miR-10b contribute to E2 release and cell apoptosis in ovarian granulosa cells by targeting CYP19A1. Mol. Cell. Endocrinol. 476 , 84–95 (2018).
Liu, J. et al . MiR-92a inhibits porcine ovarian granulosa cell apoptosis by targeting Smad7 gene. FEBS Lett. 588 , 4497–4503 (2014).
Li, X. et al . Extraction and identification of exosomes from three different sources of human ovarian granulosa cells and analysis of their differential miRNA expression profiles. J. Assist. Reprod. Genet. 41 , 1371–1385 (2024).
Wang, C. et al . Identification of Differentially Expressed mRNAs and miRNAs and Related Regulatory Networks in Cumulus Oophorus Complexes Associated with Fertilization. Reprod. Sci. 31 , 1408–1419 (2024).
The role of WNT signaling in adult ovarian folliculogenesis. Reproduction 150 (4), R137-R148, https://doi.org/10.1530/REP-14-0685 (2015).
Bahrami, A. et al . Dynamic modeling of folliculogenesis signaling pathways in the presence of miRNAs expression. Journal of Ovarian Research 10 (1), https://doi.org/10.1186/s13048-017-0371-y (2017).
Zheng, W., Nagaraju, G., Liu, Z. & Liu, K. Functional roles of the phosphatidylinositol 3-kinases (PI3Ks) signaling in the mammalian ovary. Molecular and Cellular Endocrinology 356 (1–2), 24–30, https://doi.org/10.1016/j.mce.2011.05.027 (2012).
Zhou, Q. et al . miRNA profiling of granulosa cell-derived exosomes reveals their role in promoting follicle development. J. Cell. Physiol. 239 , 20–35 (2024).
Aoki, S. et al . miRNAs in Follicular and Oviductal Fluids Support Global DNA Demethylation in Early-Stage Embryos. Int. J. Mol. Sci . 25 (2024).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37 , 907–915 (2019).
Danecek, P. et al . Twelve years of SAMtools and BCFtools. Gigascience 10 (2021).
Kovaka, S. et al . Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol . 20 (2019).
StringTie. http://ccb.jhu.edu/software/stringtie/index.shtml?t=manual .
Dhori, X. Gioiosa, S. & Gonfloni, S. Supplementary Materials. Figshare https://doi.org/10.6084/m9.figshare.26245832.v3 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq 2. Genome Biol . 15 (2014).
Wu, T. et al . clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. (Cambridge 2 (2021).
Bioconductor - org.Hs.eg.db. https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html .
Vaquero-Garcia, J. et al . RNA splicing analysis using heterogeneous and large RNA-seq datasets. Nat. Commun . 14 (2023).
Vaquero-Garcia, J. et al . A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife 5 (2016).
MAJIQ | Welcome. https://majiq.biociphers.org/ .
Csárdi, G. & Nepusz, T. The igraph software package for complex network research. (2006).
Friedländer, M. R., MacKowiak, S. D., Li, N., Chen, W. & Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40 , 37–52 (2012).
NEXTFLEX Small RNA-Seq v3 legacy - PerkinElmer Applied Genomics. https://perkinelmer-appliedgenomics.com/home/products/library-preparation-kits/small-rna-library-prep/nextflex-small-rna-seq-kit-v3-legacy/ .
CRAN - Package VennDiagram. https://cran.r-project.org/web/packages/VennDiagram/index.html .
Ru, Y. et al . The multiMiR R package and database: integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res. 42 , e133–e133 (2014).
Article ADS PubMed PubMed Central Google Scholar
Dhori, X., Gioiosa, S. & Gonfloni, S. Small RNA-seq analysis datasets. Figshare https://doi.org/10.6084/m9.figshare.23822448.v1 (2024).
Download references
Acknowledgements
We acknowledge CINECA for the availability of high-performance computing resources and specialist support. This research was funded by the University of Rome “Tor Vergata”-Progetto di Ateneo 2021.
Author information
These authors contributed equally: Silvia Gioiosa, Stefania Gonfloni.
Authors and Affiliations
CINECA, Super Computing Applications and Innovation Department, Via dei Tizii 6B, 000185, Roma, Italy
Xhulio Dhori & Silvia Gioiosa
Department of Biology, University of Roma, via della Ricerca Scientifica 00133, Roma, Italy
Xhulio Dhori & Stefania Gonfloni
You can also search for this author in PubMed Google Scholar
Contributions
S. Gio. and S. Go. conceived the study. X.D., S. Gio. and S. Go. contributed to sample preparation and metadata retrieval. X.D. and S. Gio. performed bioinformatics analysis, designed the figures, tables and supplementary materials. S. Gio. verified the analytical methods and supervised the work from a bioinformatics perspective and S. Go. from a biological point of view. All authors contributed to the interpretation of the results, provided critical feedback, and contributed to the final version of the manuscript.
Corresponding authors
Correspondence to Silvia Gioiosa or Stefania Gonfloni .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary figures, supplementary material 9, supplementary table 1, supllementary table2, supplementary table3, supplementary table4, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Dhori, X., Gioiosa, S. & Gonfloni, S. An integrated analysis of multiple datasets reveals novel gene signatures in human granulosa cells. Sci Data 11 , 972 (2024). https://doi.org/10.1038/s41597-024-03715-0
Download citation
Received : 05 January 2024
Accepted : 01 August 2024
Published : 06 September 2024
DOI : https://doi.org/10.1038/s41597-024-03715-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

COMMENTS
Abstract. Multiple regression is one of the most significant forms of regression and has a wide range. of applications. The study of the implementation of multiple regression analysis in different ...
158 PART II: BAsIc And AdvAnced RegRessIon AnAlysIs 5A.2 Statistical Regression Methods The regression procedures that we cover in this chapter are known as statistical regression methods.The most popular of these statistical methods include the standard, forward, backward, and stepwise meth- ods, although others (not covered here), such as the Mallows Cp method (e.g., Mallows, 1973) and the
The formula for a multiple linear regression is: = the predicted value of the dependent variable. = the y-intercept (value of y when all other parameters are set to 0) = the regression coefficient () of the first independent variable () (a.k.a. the effect that increasing the value of the independent variable has on the predicted y value ...
When could this happen in real life: Time series: Each sample corresponds to a different point in time. The errors for samples that are close in time are correlated. Spatial data: Each sample corresponds to a different location in space. Grouped data: Imagine a study on predicting height from weight at birth. If some of the subjects in the study are in the same family, their shared environment ...
Regression analysis is often called "ordinary least squares" (OLS) analysis because the method of determining which line best "fits" the data is to minimize the sum of the squared residuals or erros of a line put through the data. Figure 13.8. Estimated Equation: C = b0 +b1lncome + e C = b 0 + b 1 lncome + e.
With multiple regression what we're doing is looking at the effect of each variable, while holding the other variable constant. Specifically, a one unit increase in computers is associated with an increase of math scores of.002 points when holding the number of students constant, and that change is highly significant.
These questions can in principle be answered by multiple linear regression analysis. In the multiple linear regression model, Y has normal distribution with mean. The model parameters β 0 + β 1 + +β ρ and σ must be estimated from data. β 0 = intercept. β 1 β ρ = regression coefficients. σ = σ res = residual standard deviation
11.1 Matrix Algebra and Multiple Regression. Matrix algebra is widely used for the derivation of multiple regression because it permits a compact, intuitive depiction of regression analysis. For example, an estimated multiple regression model in scalar notion is expressed as: \(Y = A + BX_1 + BX_2 + BX_3 + E\).
A regression analysis is used for one (or more) of three purposes: modeling the relationship between x and y; prediction of the target variable (forecasting); and testing of hypotheses. The chapter introduces the basic multiple linear regression model, and discusses how this model can be used for these three purposes.
Multiple regression is sometimes used simply to understand factors that are relevant to predicting an outcome, but it is also used in science to help establish that the relationship between two variables is a causal one and to understand complex relationships that simply cannot be understood with bivariate analyses.
Multiple Regression. Andrew F. Siegel, in Practical Business Statistics (Sixth Edition), 2012 Separate Regressions. A different approach to multiple regression analysis of multivariate data that includes a qualitative variable is to divide up the data set according to category and then perform a separate multiple regression for each category. For example, you might have two analyses: one for ...
2.1 Introduction to Multiple Regression. Bivariate, or simple, regression examines the effect of an independent variable (X) on the dependent variable (Y). Multiple regression extends this idea by con-sidering the effects of multiple independent variables (X's) on the dependent variable (Y). It is almost always more realistic for there to be ...
Second, multiple regression is an extraordinarily versatile calculation, underly-ing many widely used Statistics methods. A sound understanding of the multiple regression model will help you to understand these other applications. Third, multiple regression offers our first glimpse into statistical models that use more than two quantitative ...
The multiple linear regression is the most widely used multivariate technique in non-laboratory sciences such as social sciences for examining the assumed causal relationships between a set of independent variables and a dependent variable. The dependent variable is a phenomenon which we seek to explain. And an independent variable is a phenomenon which we assume as a causal factor, a thing ...
The objective of multiple regression analysis is to describe the influence of a set of independent variables \(X_{j}\) on a dependent variable \(Y\) and to determine what happens to \(Y\) when a variable \(X_{j}\) changes or is changed. The difference to linear simple regression is that we are not only examing only one independent variable, but ...
The study of the implementation of multiple regression analysis in different settings contributes to the development of relevant theories and the improvement of models. In this paper,
When we use the regression sum of squares, SSR = Σ ( ŷi − Y−) 2, the ratio R2 = SSR/ (SSR + SSE) is the amount of variation explained by the regression model and in multiple regression is ...
Regression analysis is a quantitative research method which is used when the study involves modelling and analysing several variables, where the relationship includes a dependent variable and one or more independent variables. In simple terms, regression analysis is a quantitative method used to test the nature of relationships between a dependent variable and one or more independent variables.
Regression analysis is an important ... one can perform a multivariable linear regression to study the effect or association with multiple ... Department of Health Research Methods, Evidence ...
Introduction. This is the second part of our discussion about general linear models. In this chapter we extend linear regression from one to many predictor variables.We also introduce logistic regression, which uses logistic function to model binary outcome variables.Extensions to address ordinal outcome variables are also presented. We conclude with a discussion of model selection, which ...
Multiple and Curvilinear Regression. R.H. Riffenburgh, in Statistics in Medicine (Third Edition), 2012 Visualizing the Models. The concept of multiple regression is similar to that of simpler regression, in that the dependent variable is related to the independent variables by a best fit. However, the geometry is extended from a line in x,y dimensions to a plane in x 1,x 2,y dimensions or to ...
Consequently, and similar to quantitative traditions in sister-disciplines such as education and psychology (see Skidmore & Thompson, 2010), second language researchers have turned increasingly to multiple regression. The present study employs research synthetic techniques to describe and evaluate the use of this procedure in the field.
Multiple regression analysis is a method that analysts and statisticians use to understand and create conclusions about multiple regression. In this article, we offer a multiple regression analysis definition, list the formula for calculating multiple regression and explain how to calculate multiple regression with an example to provide more ...
Multiple regression is an extension of simple linear regression. It is used when we want to predict the value of a variable based on the value of two or more other variables. The variable we want to predict is called the dependent variable (or sometimes, the outcome, target or criterion variable). The variables we are using to predict the value ...
Quantitative Research Methods. Correlation is the relationship or association between two variables. There are multiple ways to measure correlation, but the most common is Pearson's correlation coefficient (r), which tells you the strength of the linear relationship between two variables. The value of r has a range of -1 to 1 (0 indicates no ...
In this article, we focus on a potentially more straightforward issue: analyses of randomized experiments. Much has been written about the analysis of randomized trials (see Shadish et al., 2002).In the ideal case, a study is planned, participants are assigned randomly to treatments, and the analyses follow a preregistered plan to avoid the possible biases that may be introduced if analyses ...
Multiple regression analyses revealed that older students and females exhibited more favorable attitude compared to their younger and male counterparts, respectively.
To address the research questions concerning the fundamental mechanisms influencing the relationship between individuals' knowledge of vaccines and eHealth literacy, a survey targeting Chinese ...
Discover the world's research. ... An ensemble learning method that constructs multiple decision trees and ... metrics useful for binary classification or threshold-based analysis in regression .
The p-values were adjusted for multiple testing using the appropriate method Benjamini-Hochberg correction method. The results of the GO enrichment analysis were visualized using various plotting ...